Effortless Clinical Text Analysis with Advanced Pretrained Pipelines
This blog post explores Healthcare NLP’s Task-Based Clinical Pretrained Pipelines, showcasing how they streamline clinical text analysis with just one-liner codes. By demonstrating the explain_clinical_doc_granular pipeline in a real-world scenario, we illustrate its capabilities in Named Entity Recognition (NER), Assertion Status, and Relation Extraction. These pipelines provide an efficient way to extract medical insights from unstructured clinical text, offering valuable tools for healthcare professionals and researchers.
Clinical text is a treasure trove of patient information, but extracting actionable insights can be both complex and time-consuming. Traditional methods demand significant data preprocessing, advanced models, and specialized domain expertise. However, with Healthcare NLP’s task-based pretrained pipelines, these challenges can be overcome with simple one-liner solutions that tackle everything from entity recognition to de-identification.
As clinical data volumes grow, the demand for quick, reliable, and efficient analysis tools intensifies. Healthcare NLP’s pretrained pipelines empower professionals to extract valuable information from unstructured medical texts — including clinical notes, pathology reports, and health records — using a few simple commands. This automation streamlines decision-making, reduces manual effort, and ultimately enhances patient care efficiency.
Traditionally, managing health records has been a labor-intensive process, but Natural Language Processing (NLP) now offers solutions to automate, partially or entirely, this task, enabling healthcare providers to analyze vast amounts of data in real-time.
What Is a Pipeline?
In machine learning, a pipeline is a structured workflow that applies a series of algorithms in a defined sequence, passing the results from one step to the next. This workflow, widely used in Apache Spark ML, ensures smooth data flow and optimized performance. Similarly, Healthcare NLP pipelines follow this principle, enabling seamless text processing for clinical applications.
Each step relies on a combination of Transformers and Estimators, working together as an integrated system. This synergy simplifies complex text analysis tasks, making Healthcare NLP an invaluable tool for efficient and accurate data processing.
The Power of Task-Based Pipelines
With Healthcare NLP pipelines, healthcare providers can rapidly extract key clinical information, determine assertion status (whether a condition is present, hypothetical, or absent), and map concepts to standardized medical codes (ICD, RxNorm, SNOMED CT). This automation accelerates clinical decision-making, aiding in better management of health records.
By leveraging pretrained pipelines, professionals can process clinical text faster, extract actionable insights with minimal effort, and focus on improving patient outcomes — all with just a few lines of code.
Emerging Trends in Clinical NLP
A major breakthrough shaping the current landscape of clinical NLP is the shift toward context-aware, multimodal processing. Healthcare NLP systems are no longer limited to textual data alone, they are increasingly designed to integrate clinical text with imaging, laboratory results, genomics, and sensor data. This convergence allows models to provide a richer and more precise understanding of patient status. For instance, task-based NLP pipelines are now capable of aligning clinical narratives with imaging findings or lab markers, leading to more accurate condition tracking and risk stratification. These advancements are already improving clinical workflows in diagnostics and treatment planning.
Another defining trend is the rapid expansion of real-time NLP applications within clinical settings. Many health systems have started embedding task-based NLP pipelines directly into EHR interfaces and clinical decision support tools. This integration enables instant summarization of medical histories, detection of emerging conditions, and context-based recommendations at the point of care. Recent deployments show measurable reductions in documentation time and error rates, making NLP not just a backend analytics tool but an active component of clinical decision-making and operational efficiency.
Finally, federated and privacy-preserving NLP approaches are becoming the backbone of large-scale healthcare innovation. Instead of centralizing sensitive patient data, organizations are adopting decentralized training and fine-tuning strategies that allow models to learn from distributed clinical datasets without compromising privacy. This shift aligns with regulatory priorities and accelerates the deployment of domain-specific models across institutions. As a result, healthcare NLP systems are becoming more robust, generalizable, and trustworthy, fueling innovation while maintaining compliance and patient trust.
Introducing Healthcare NLP & LLM
The Healthcare NLP Library is a powerful component of John Snow Labs’ Healthcare NLP platform, designed to streamline natural language processing (NLP) tasks in the healthcare domain. With over 2,500 pre-trained models and pipelines, this library empowers professionals to efficiently extract critical medical information, perform Named Entity Recognition (NER) for clinical concepts, and analyze complex medical text. Regularly updated with cutting-edge algorithms, the library enables seamless processing of unstructured medical data from electronic health records (EHRs), clinical notes, and biomedical literature, transforming raw text into valuable insights.
Custom Large Language Models for Healthcare
John Snow Labs has developed specialized Large Language Models (LLMs) tailored for diverse healthcare applications. These models come in various sizes and quantization levels, enabling tasks such as:
- Summarizing medical notes
- Answering clinical questions
- Performing Retrieval-Augmented Generation (RAG)
- Recognizing medical entities with NER
- Enabling healthcare-related conversational AI
By integrating domain-specific knowledge with state-of-the-art NLP techniques, these LLMs enhance clinical decision-making, automate documentation, and support advanced medical research.
Resources & Learning Opportunities
- GitHub Repository: John Snow Labs’ GitHub repository is a collaborative hub where users can access open-source code, tutorials, and projects to further their expertise in Healthcare NLP.
- Certification Training: John Snow Labs offers certification programs to help users master the Healthcare NLP Library, with structured learning paths guided by industry experts.
- Live Demos & Interactive Testing: The John Snow Labs Demo Page allows users to explore the library’s capabilities and interact with models, offering a hands-on experience to better understand its real-world applications in healthcare and beyond.
- Models Hub: John Snow Labs’ Models Hub provides state-of-the-art NLP and LLM models for Open-source, Healthcare applications, offering pre-trained solutions for various tasks.
Task Based Pretrained Pipelines
John Snow Labs provides a range of task-specific pre-trained pipelines to streamline clinical text processing. Below is an overview of some key Healthcare NLP pipelines, each designed to extract, analyze, and structure medical information efficiently.
- Explain Clinical Doc Generic This pipeline is designed to extract all clinical/medical entities, assign assertion status to the extracted entities, and establish relations between the extracted entities from the clinical texts.
- Explain Clinical Doc Granular This pipeline is designed to extract all clinical/medical entities, assign assertion status to the extracted entities, and establish relations between the extracted entities from the clinical texts.
- Explain Clinical Doc Biomarker This specialized biomarker pipeline can extract biomarker entities, classify sentences whether they contain biomarker entities or not, and establish relations between the extracted biomarker and biomarker results from the clinical documents.
- Explain Clinical Doc Oncology This specialized oncology pipeline can extract oncological entities, assign assertion status to the extracted entities, and establish relations between the extracted entities from the clinical documents.
- Explain Clinical Doc Radiology This pipeline is designed to extract all clinical/medical entities, assign assertion status to the extracted entities, and establish relations between the extracted entities from the clinical texts.
- Explain Clinical Doc VOP This pipeline is designed to extract healthcare-related terms entities, assign assertion status to the extracted entities, and establish relations between the extracted entities from the documents transferred from the patient’s sentences.
- Explain Clinical Doc CARP A pipeline with ner_clinical, assertion_dl, re_clinical, and ner_posology. It extracts clinical and medication entities, assigns assertion status, and finds relationships between clinical entities.
- Explain Clinical Doc ERA A pipeline with ner_clinical_events, assertion_dl, and re_temporal_events_clinical. It extracts clinical entities, assigns assertion status, and finds temporal relationships between clinical entities.
- Explain Clinical Doc ADE A pipeline for Adverse Drug Events (ADE) with ner_ade_biobert, assertion_dl_biobert, classifierdl_ade_conversational_biobert, and re_ade_biobert. It classifies the document, extracts ADE and DRUG clinical entities, assigns assertion status to ADE entities, and relates Drugs with their ADEs.
- Explain Clinical Doc Medication A pipeline for detecting posology entities with the ner_posology_large NER model, assigning their assertion status with assertion_jsl model, and extracting relations between posology-related terminology with posology_re relation extraction model.
- Explain Clinical Doc Risk Factors This pipeline is designed to extract all clinical/medical entities, which may be considered as risk factors from text, assign assertion status to the extracted entities, and establish relations between the extracted entities.
- Explain Clinical Doc Public Health This specialized public health pipeline extracts public health-related entities, assigns assertion status to the extracted entities, and establishes relations between the extracted entities from the clinical documents. In this pipeline, five NER, one assertion, and one relation extraction model were used to achieve those tasks.
- Explain Clinical Doc SDOH This pipeline is designed to extract all clinical/medical entities, assertion status, and relation information, which may be considered as Social Determinants of Health (SDOH) entities from text.
- Explain Clinical Doc Mental Health This pipeline is designed to extract all mental health-related entities, assertion status, and relation information from text.
- NER Medication Generic Pipeline This pre-trained pipeline is designed to identify generic DRUG entities in clinical texts. It was built on top of the ner_posology_greedy, ner_jsl_greedy, ner_drugs_large, and drug_matcher models to detect the entities DRUG, DOSAGE, ROUTE, and STRENGTH, chunking them into a larger entity as DRUG when they appear together.
Using a Pretrained Pipeline
John Snow Labs’ Healthcare NLP provides ready-to-use pre-trained pipelines to extract valuable insights from clinical text effortlessly. You can load and use the explain_clinical_doc_granular pipeline with just a few lines of code.
This pipeline is designed to:
- Extract all clinical/medical entities from clinical texts.
- Assign assertion status to the extracted entities, indicating whether they are confirmed, negated, or hypothetical.
- Establish relations between the extracted entities to provide a deeper understanding of the clinical context.
With the explain_clinical_doc_granular pipeline, you can automatically process clinical documents to uncover essential details about patient conditions, treatments, and more, all while ensuring high precision and accuracy. Now, let’s load and call the pipeline using the following code:
from sparknlp.pretrained import PretrainedPipeline
pipeline = nlp.PretrainedPipeline("explain_clinical_doc_granular", "en", "clinical/models")
Consider the following clinical note from a physician documenting a patient’s condition:
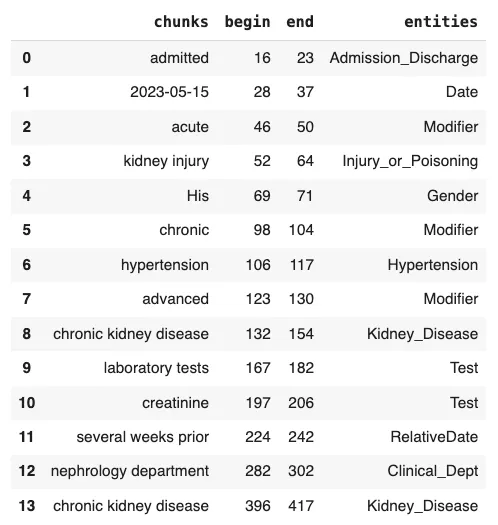
“The patient was admitted on 2023–05–15 due to acute kidney injury.His medical history includes chronic hypertension and advanced chronic kidney disease.Earlier laboratory tests had detected creatinine levels assessed several weeks prior.The patient has been referred to the nephrology department for further evaluation.The patient’s family history includes both parents diagnosed with chronic kidney disease.”
text = """The patient was admitted on 2023-05-15 due to acute kidney injury. His medical history includes chronic hypertension and advanced chronic kidney disease. Earlier laboratory tests had detected creatinine levels assessed several weeks prior. The patient has been referred to the nephrology department for further evaluation. The patient's family history includes both parents diagnosed with chronic kidney disease. """ result = pipeline.fullAnnotate(text)[0]
After processing our sample medical text with the pretrained pipeline, we will extract and present the Named Entity Recognition (NER), Assertion Status, and Relation Extraction results.
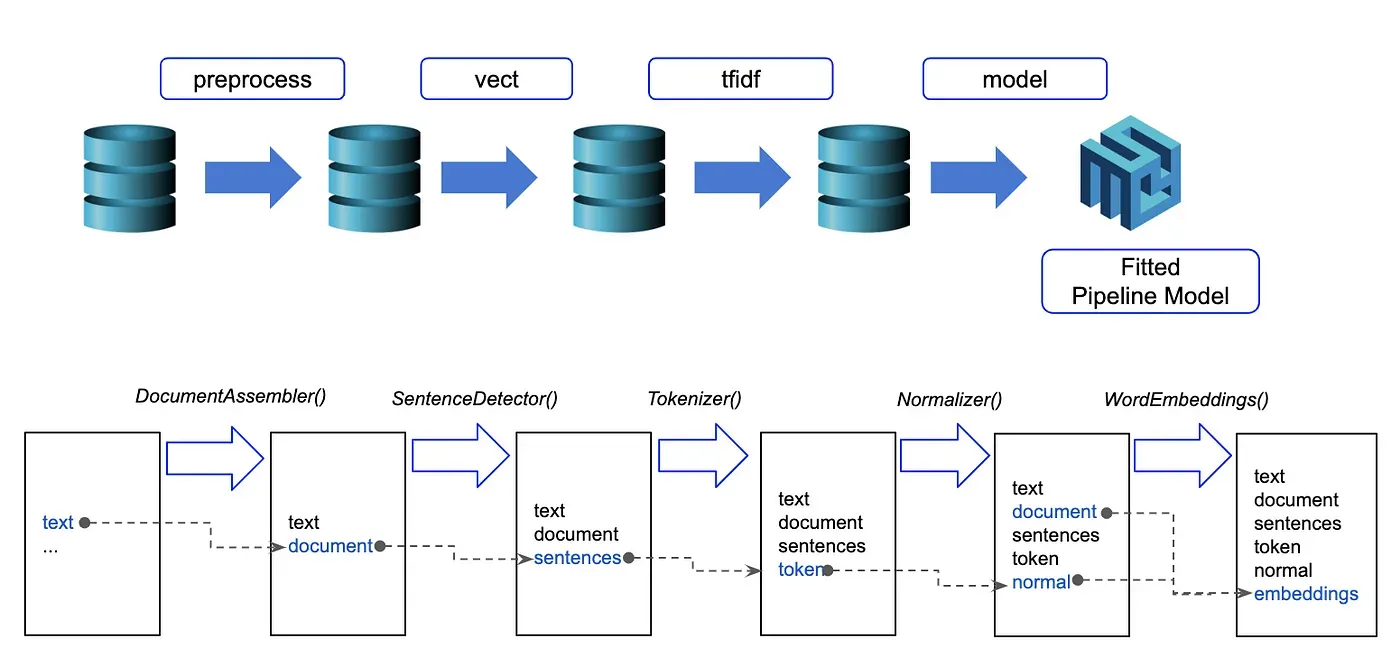
Extracting Named Entities
Once you have processed clinical text using a pre-trained Healthcare NLP pipeline, you can extract and visualize Named Entity Recognition (NER) results with the following code:
import pandas as pd
chunks=[]
entities=[]
begins=[]
ends=[]
for n in result['jsl_ner_chunk']:
chunks.append(n.result)
begins.append(n.begin)
ends.append(n.end)
entities.append(n.metadata['entity'])
df = pd.DataFrame({'chunks':chunks, 'begin':begins, 'end':ends, 'entities':entities})
df
NER Results
Visualization of NER Results
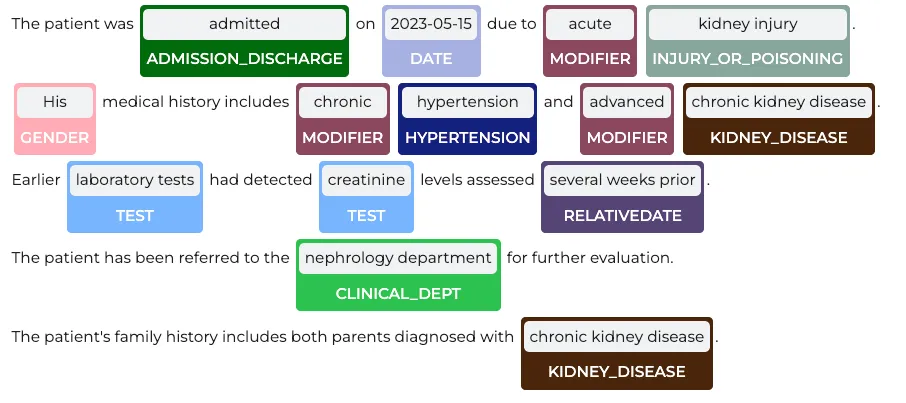
Extracting Assertion Status
In clinical NLP, assertion status helps determine whether an extracted medical entity is present, absent, planned, family, past, hypothetical, possible, someoneelse, within the text. Using the Healthcare NLP library, we can extract assertion status for named entities with the following code:(We do not check the assertion for every entity . Specifically, we exclude entities such as ‘Admission_Discharge,’ ‘Clinical_Dept,’ ‘Gender,’ ‘Date,’ and ‘ADMISSION_DISC.’ from assertion analysis.)
import pandas as pd
chunks = []
entities = []
status = []
begin = []
end = []
for n, m in zip(result['assertion_ner_chunk'], result['assertion']):
chunks.append(n.result)
begin.append(n.begin)
end.append(n.end)
entities.append(n.metadata['entity'])
status.append(m.result)
df = pd.DataFrame({'chunks': chunks, 'begin': begin, 'end': end, 'entities': entities, 'assertion': status})
df
Assertion Status Results
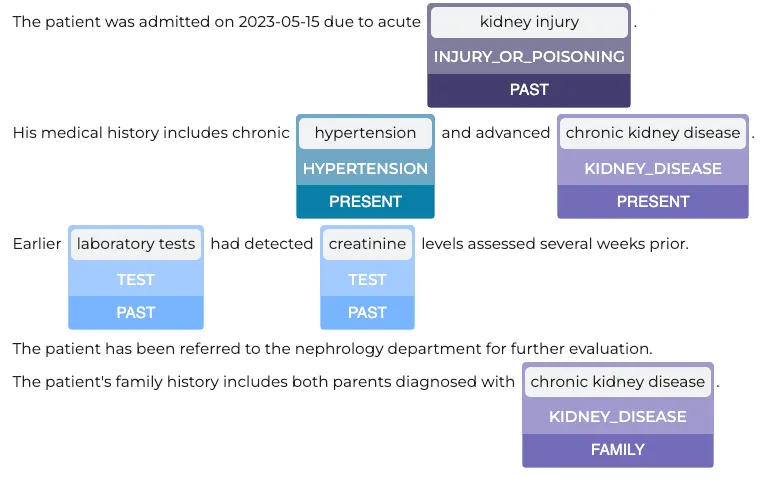
Visualization of Assertion Status Results
Extracting Relations Between Medical Entities
In clinical NLP, relation extraction helps identify meaningful connections between medical entities, such as:
- is_finding_of → A symptom or condition is linked to a diagnosis
- is_date_of → A specific date corresponds to an event (e.g., diagnosis date)
Using John Snow Labs’ Healthcare NLP, we can extract relations with the following code:
annotations = pipeline.fullAnnotate(text) rel_df = get_relations_df(annotations, 'all_relations') rel_df[rel_df.relation != "O"]
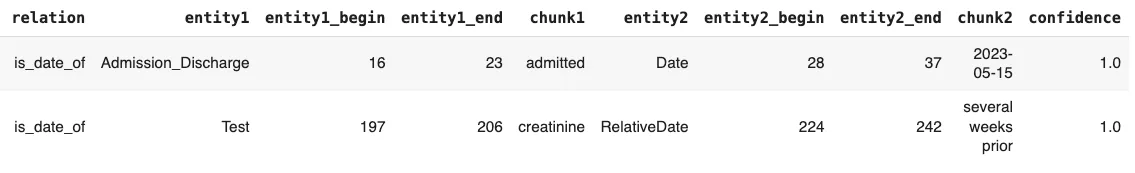
Relation Extraction Results
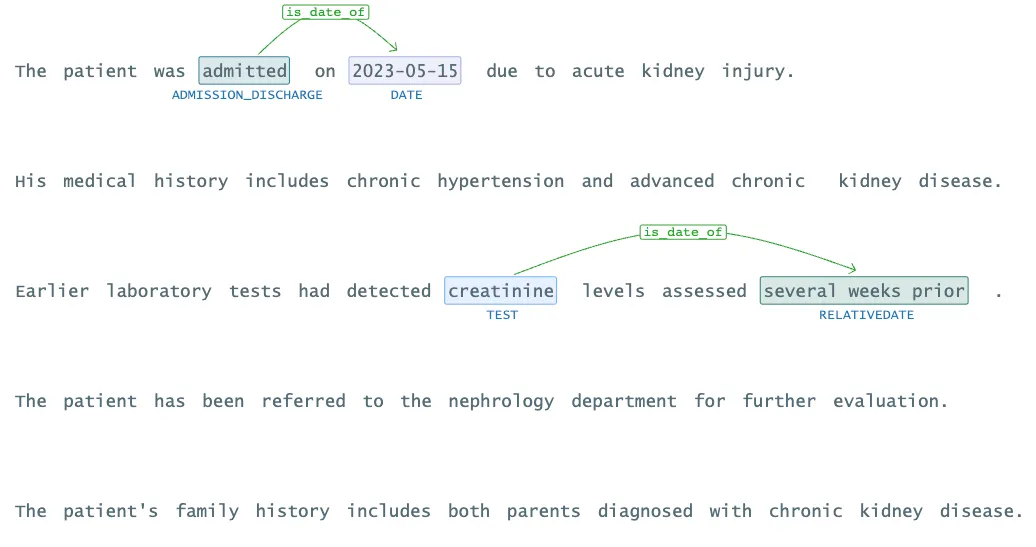
Visualization of Relation Extraction Results
PipelineTracer and PipelineOutputParser
The PipelineTracer class is a powerful and flexible tool that tracks every stage of a pipeline, providing detailed insights into entities, assertions, de-identification, classification, and relationships. It also plays a key role in building parser dictionaries for creating a PipelineOutputParser.
This class enables users to print the pipeline schema, generate parser dictionaries, and retrieve possible assertions, relationships, and entities. Additionally, it offers seamless access to parser dictionaries and existing pipeline diagrams, making it an essential component for pipeline analysis and debugging.
The following code demonstrates how to utilize PipelineTracer and PipelineOutputParser to explore our pipeline’s structure. This provides an overview of the components used in the pipeline, helping to refine and adapt it for specific tasks.
PipelineTracer:
tracer = PipelineTracer(pipeline)
print("Entities: ", tracer.getPossibleEntities())
print("Assertions: ", tracer.getPossibleAssertions())
print("Relations: ", tracer.getPossibleRelations())
Output:
Entities: ['Injury_or_Poisoning', 'Direction', 'Test', 'Route' 'Admission_Discharge', 'Death_Entity', 'Oxygen_Therapy', 'Relationship_Status' 'Drug_BrandName', 'Duration', 'Alcohol', 'Triglycerides' 'Date', 'Hyperlipidemia', 'Respiration', 'Birth_Entity' 'VS_Finding', 'Age', 'Vaccine_Name', 'Social_History_Header' 'Labour_Delivery', 'Medical_Device', 'Family_History_Header', 'BMI' 'Fetus_NewBorn', 'Temperature', 'Section_Header', 'Communicable_Disease' 'ImagingFindings', 'Psychological_Condition', 'Obesity', 'Sexually_Active_or_Sexual_Orientation' 'Modifier', 'Vaccine', 'Symptom', 'Pulse' 'Kidney_Disease', 'Oncological', 'EKG_Findings', 'Medical_History_Header' 'Cerebrovascular_Disease', 'Blood_Pressure', 'Diabetes', 'O2_Saturation' 'Heart_Disease', 'Frequency', 'Employment', 'Disease_Syndrome_Disorder' 'Pregnancy', 'RelativeDate', 'Procedure', 'Race_Ethnicity' 'Hypertension', 'External_body_part_or_region', 'Imaging_Technique', 'Test_Result' 'Substance', 'Treatment', 'Clinical_Dept', 'Drug_Ingredient' 'LDL', 'Diet', 'Substance_Quantity', 'Allergen' 'Gender', 'RelativeTime', 'Total_Cholesterol', 'Internal_organ_or_component' 'Vital_Signs_Header', 'Height', 'Smoking', 'Form' 'Strength', 'Weight', 'Time', 'Dosage' 'Overweight', 'HDL'] Assertions: ['Family', 'Past', 'Hypothetical', 'Possible', 'SomeoneElse', 'Planned', 'Absent', 'Present'] Relations ['is_finding_of', 'is_result_of', 'is_date_of']
PipelineOutputParser:
light_result= pipeline.fullAnnotate(text) pipeline_parser = PipelineOutputParser(column_maps) result_parser = pipeline_parser.run(light_result) result_parser['result'][0]
Output:
{'document_identifier': 'explain_clinical_doc_granular',
'document_id': 0,
'document_text': ["The patient was admitted on 2023-05-15 due to acute kidney injury. \nHis medical history includes chronic hypertension and advanced chronic kidney disease. \nEarlier laboratory tests had detected creatinine levels assessed several weeks prior.\nThe patient has been referred to the nephrology department for further evaluation. \nThe patient's family history includes both parents diagnosed with chronic kidney disease.\n"],
'entities': [{'chunk_id': '79a7e38f',
'chunk': 'admitted',
'begin': 16,
'end': 23,
'ner_label': 'Admission_Discharge',
'ner_source': 'jsl_ner_chunk',
'ner_confidence': '0.9992'},
{'chunk_id': 'd3a6861a',
'chunk': '2023-05-15',
'begin': 28,
'end': 37,
'ner_label': 'Date',
'ner_source': 'jsl_ner_chunk',
'ner_confidence': '0.4348'},
{'chunk_id': 'a1de0526',
'chunk': 'acute',
'begin': 46,
'end': 50,
'ner_label': 'Modifier',
'ner_source': 'jsl_ner_chunk',
'ner_confidence': '0.9388'},
...
For more details on PipelineTracer and PipelineOutputParser, Please refer to the official notebook from John Snow Labs.
Customizing Pretrained Pipelines in Healthcare NLP
Healthcare NLP provides the flexibility to customize pretrained pipelines according to specific use cases, allowing users to modify, add, or remove stages as needed. This capability ensures that entity extraction, assertion detection, relation identification, deidentification align with the requirements of different medical applications. For a detailed guide on how to customize pretrained pipelines, refer to the Customization of Pretrained Pipelines notebook:
🔗 Customize Your Pretrained Pipeline
This resource walks through modifying pipeline components, ensuring optimal performance for specialized NLP tasks in healthcare.
Conclusion
Task-based clinical NLP revolutionizes the way we extract insights from medical text. With just a single line of code, users can perform entity recognition, assertion detection, and relation extraction, transforming unstructured clinical notes into structured, actionable data. By leveraging pre-trained models in Healthcare NLP, medical professionals, researchers, and data scientists can accelerate clinical decision-making, enhance patient care, and unlock new opportunities in healthcare AI.
Moreover, Healthcare NLP offers a wide range of specialized pipelines tailored for various applications, including entity deidentification and resolver pipelines that map clinical entities to standardized codes such as SNOMED CT, ICD-10, and RXNORM. These pipelines can be customized to fit specific use cases, ensuring flexibility and adaptability for different healthcare and research needs. Whether it’s structuring patient records, supporting clinical trials, or improving electronic health records (EHR) systems, Healthcare NLP provides scalable, efficient solutions that empower users to harness the full potential of AI in medicine.
FAQ
What is a “one-liner” clinical NLP pipeline and when should I use it?
A one-liner pipeline is a prebuilt workflow that chains NER, assertion status, and relation extraction so you can analyze clinical text with a single call, ideal for proofs of concept, rapid dashboards, and bulk processing without model tuning. Use it when you need reliable, domain-specific extraction fast, and switch to customization only if you have very specific entities, ontologies, or UI/latency requirements.
What exactly does explain_clinical_doc_granular return?
It identifies clinical entities (problems, tests, treatments, etc.), assigns assertion status (present/absent/planned/family/past/hypothetical/possible/someone-else), and extracts relations (e.g., is_finding_of, is_result_of, is_date_of). You’ll get structured outputs you can turn into tables, chunks with spans and labels, assertion values per chunk, and relation triplets to connect entities.
Can I customize the pipeline (e.g., add de-identification, resolvers, or domain-specific NER)?
Yes. You can fork the prebuilt graph and add/remove stages, e.g., insert de-identification before persistence, attach RxNorm/SNOMED/ICD resolvers, or swap in specialty NER (oncology, radiology, ADE, SDOH, mental health). Keep the one-liner for quick wins, then evolve it into a tailored pipeline when you need custom labels, rules, or post-processing.
Is it suitable for real-time EHR workflows and how is PHI protected?
Pipelines can run in batch (Spark clusters) or near-real-time (Spark local, micro-services, or streaming connectors) and are often embedded in CDS/EHR components for instant summarization and alerts. PHI is protected by running fully on your infrastructure, applying de-identification before logging/analytics, enforcing role-based access, and, when training/fine-tuning, using privacy-preserving strategies (e.g., federated or on-prem training) to comply with HIPAA/GDPR and internal policies.