Spark NLP for Healthcare 2.7.3 is available now! If you’re an existing customer, expect an email with upgrade instructions.
In this release we introduce the following new models, features, and improvements:
- Massive improvements & feature enhancements in De-Identification module
- A brand-new RelationExtractionDL Annotator
- Drug Normalizer
- Confidence Scores in assertion output
- Cosine similarity metrics in entity resolvers
- AuxLabel in the metadata of entity resolvers
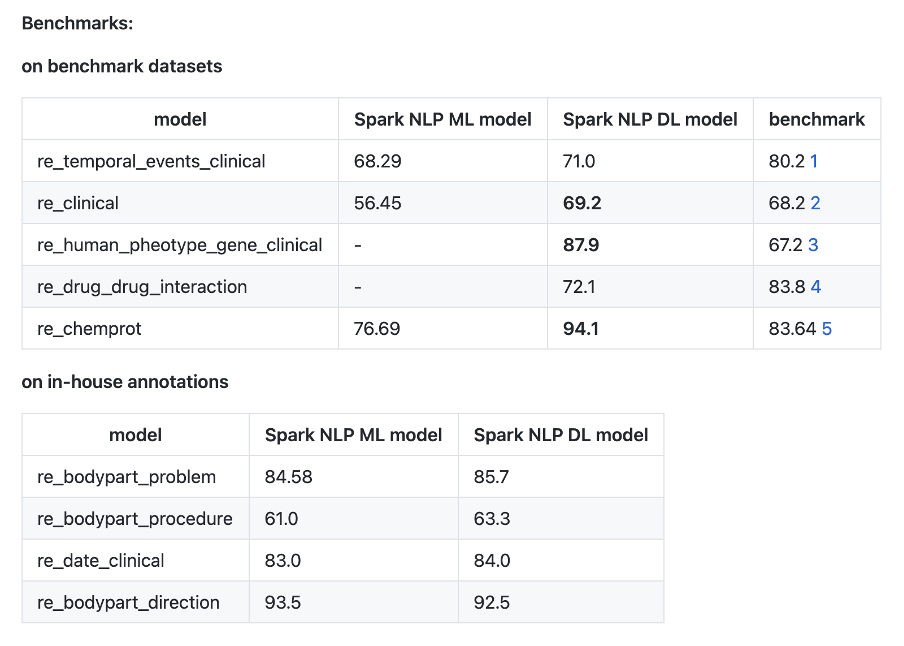
- New Relation Extraction models
- New Entity Resolver models
- New Clinical Pretrained NER model
- Bug fixes & general improvements
- Matching the version with Spark NLP open-source v2.7.3
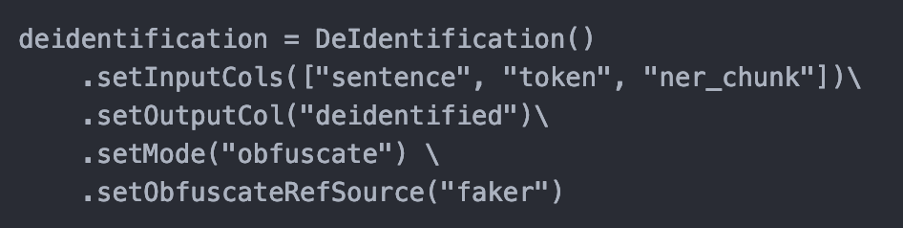
Improvements in De-Identification Module
Faker Library
This release introduces the ‘faker’ library in De-Identification Module to automatically generate random data like names, dates, addresses etc. so users do not have to specify dummy data (custom obfuscation files can still be used). It also improves the obfuscation results due to a bigger pool of random values.
How to use
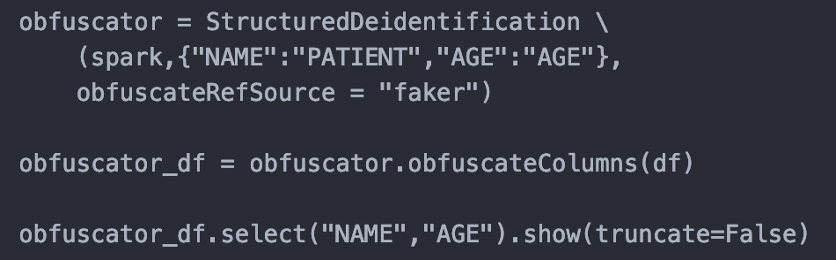
Structured De-Identification Module
This release comes with a new annotator to handle the de-identification of structured data. It allows users to define a mapping of columns and their obfuscation policy. Users can also provide dummy data and map them to columns they want to replace values in.
How to use
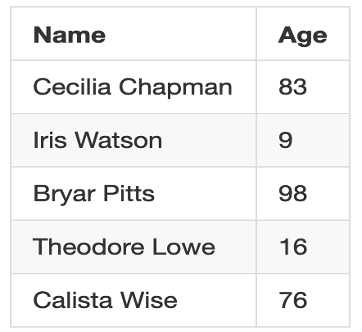
Input:
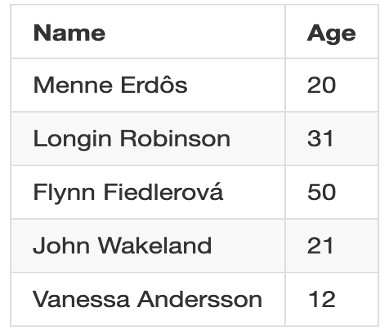
Deidentified:
Introducing SOTA Relation Extraction model using BioBert
We released a brand-new end-to-end trained BERT model, resulting in massive improvements. Another new annotator (ReChunksFilter) is also developed for this new model to allow syntactic features to work well with BioBert to extract relations.
How to use
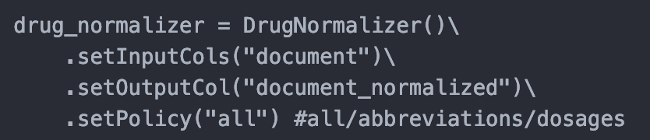
Drug Normalizer
This release comes with Drug Normalizer to standardize units of drugs and handle abbreviations in raw text or drug chunks identified by any NER model. This normalization significantly improves the performance of entity resolvers.
How to use
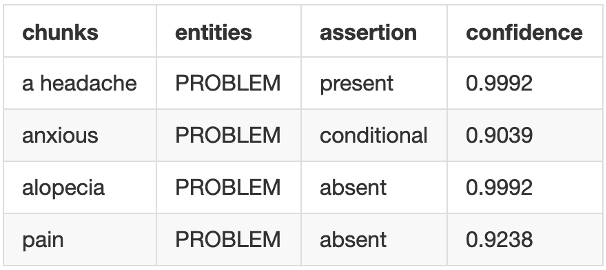
Assertion models to support confidence in the output
Just like NER output, now assertion models also provide confidence scores for each prediction.
New Relation Extraction Models
This release introduces 3 new relation extraction models between body parts and clinical entities and 1 new relation extraction model between date and clinical entities. These models are trained using a binary relation extraction approach for better accuracy.
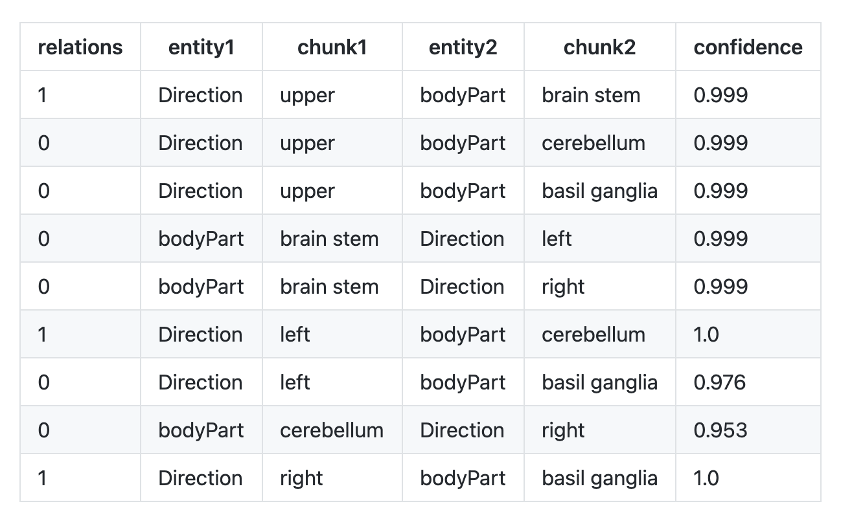
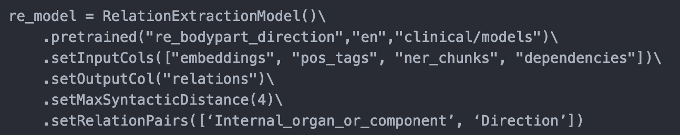
- re_bodypart_direction : Relation Extraction between Body Part and Direction entities.
Example:
Text: “MRI demonstrated infarction in the upper brain stem, left cerebellum and right basil ganglia”
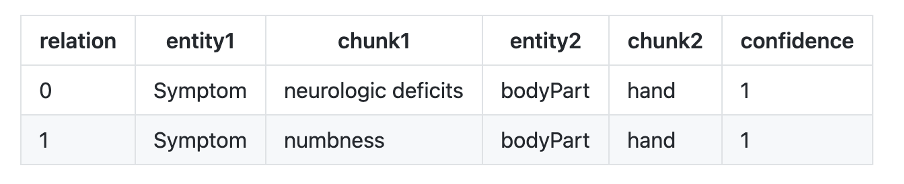
- re_bodypart_problem : Relation Extraction between Body Part and Problem entities.
Example:
Text: “No neurologic deficits other than some numbness in his left hand.”
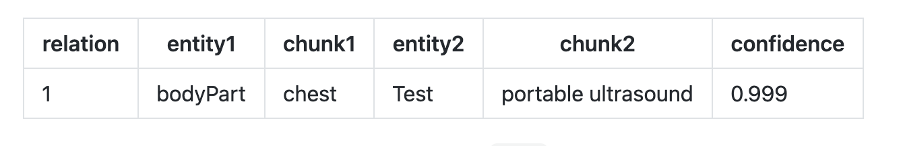
- re_bodypart_proceduretest : Relation Extraction between Body Part and Procedure, Test entities.
Example:
Text: “TECHNIQUE IN DETAIL: After informed consent was obtained from the patient and his mother, the chest was scanned with portable ultrasound.”
- re_date_clinical : Relation Extraction between Date and different clinical entities.
Example:
Text: “This 73 y/o patient had CT on 1/12/95, with progressive memory and cognitive decline since 8/11/94.”
How to use
New Resolver Models using JSL SBERT
- sbiobertresolve_icd10cm_augmented
- sbiobertresolve_cpt_augmented
- sbiobertresolve_cpt_procedures_augmented
- sbiobertresolve_icd10cm_augmented_billable_hcc
- sbiobertresolve_hcc_augmented
Returning auxiliary columns mapped to resolutions: Chunk entity resolver and sentence entity resolver now returns auxiliary data that is mapped the resolutions during training. This will allow users to get multiple resolutions with a single model without using any other annotator in the pipeline (In order to get billable codes otherwise there need to be other modules in the same pipeline)
We also added a cosine similarity metric in metadata to resolve entities and find the closest matches, resulting in better, more semantically correct results.
- sbiobertresolve_icd10cm_augmented_billable_hcc
Example:
Input Text: “bladder cancer”
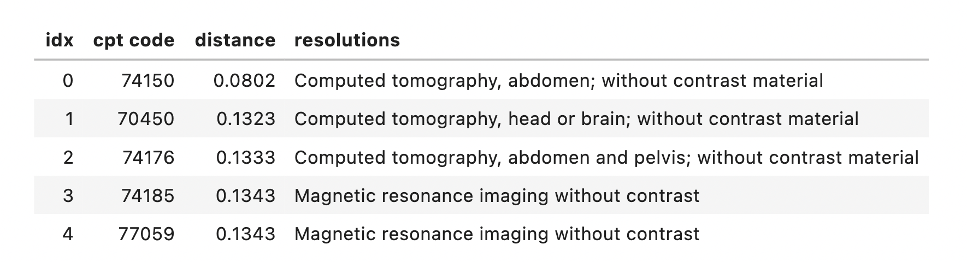
- sbiobertresolve_cpt_augmented
Example:
Input Text: “ct abdomen without contrast”
New Pretrained Clinical NER Model
- NER Radiology
Input Text: “Bilateral breast ultrasound was subsequently performed, which demonstrated an ovoid mass measuring approximately 0.5 x 0.5 x 0.4 cm in diameter located within the anteromedial aspect of the left shoulder. This mass demonstrates isoechoic echotexture to the adjacent muscle, with no evidence of internal color flow. This may represent benign fibrous tissue or a lipoma.”