We are thrilled to announce that Spark NLP for Healthcare 2.6.0 has been released.
Start your free trial – or expect an email with upgrade details if you’re already a subscriber.
Clinical Relation Extraction
This release comes with a new, deep-learning based, and trainable algorithm for relation extraction. Five pre-trained models are bundled:
re_temporal_events_clinical
re_temporal_events_enriched_clinical
re_human_phenotype_gene_clinical
re_drug_drug_interaction_clinical
re_chemprot_clinical
This Python Colab notebook shows these models in action – from detecting temporal relationships between clinical events to drug-drug interactions.
Here are a few examples of clinical relationships the temporal events model can extract:

Named Entity Recognition for Genes, Phenotypes, Chemicals, and Proteins
This release comes with three new pre-trained biomedical NER models:
ner_human_phenotype_gene_clinical
ner_human_phenotype_go_clinical
ner_chemprot_clinical
As one example, ner_chemprot_clinical when partnered with re_chemprot_clinical is useful in automatically detecting the interactions between chemicals and proteins –a crucial task in precision medicine, drug discovery, and basic clinical research. Currently, PubMed contains >28 million articles, and its annual growth rate is more than a million articles each year. A large amount of valuable chemical–protein interactions (CPIs) are hidden in the biomedical literature. Since manually extracting biomedical relations such as protein–protein interactions (PPI) and drug–drug interactions (DDI) is costly and time-consuming, some computational methods have been proposed to automate it.
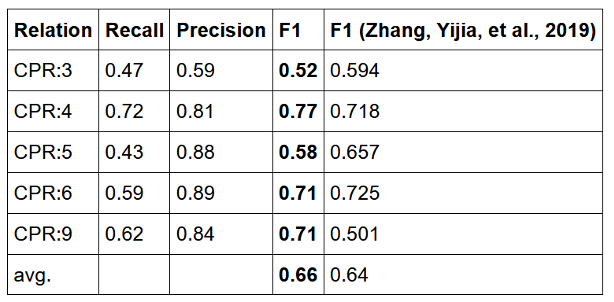
On top of the pre-trained named entity recognition model for chemicals and proteins, the pre-trained relation extraction model currently outperforms on average the most recent academic results by Zhang et. al. 2019, when trained & measured against the same benchmark:
Clinical & Legal Named Entity Recognition in German
The first time ever, we are releasing 3 licensed German models for healthcare and Legal domains. Here are the models, with links to Colab notebooks that show them in action:
– German Clinical NER model for 19 clinical entities
– German Legal NER model for 19 legal entities
Here is a visual sneak peek from inside these notebooks:
German Clinical NER

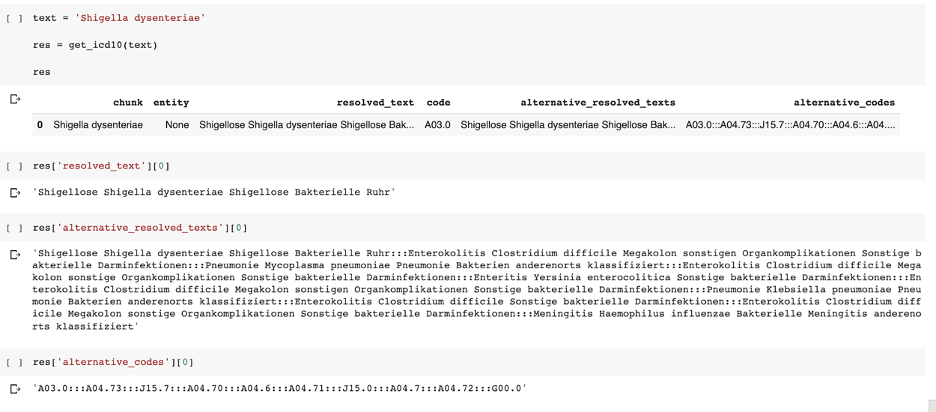
German ICD10-GM
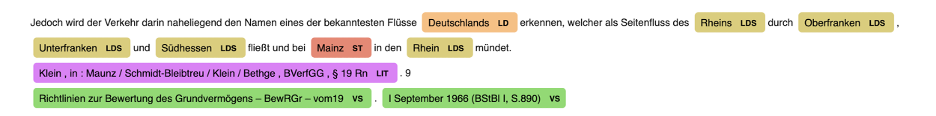
German Legal NER
Pretrained Pipelines:
The first time ever, we release three pretrained clinical pipelines to save you from building pipelines from scratch. Pretrained pipelines are already fitted using certain annotators and transformers according to various use cases and you can use them as easy as follows:
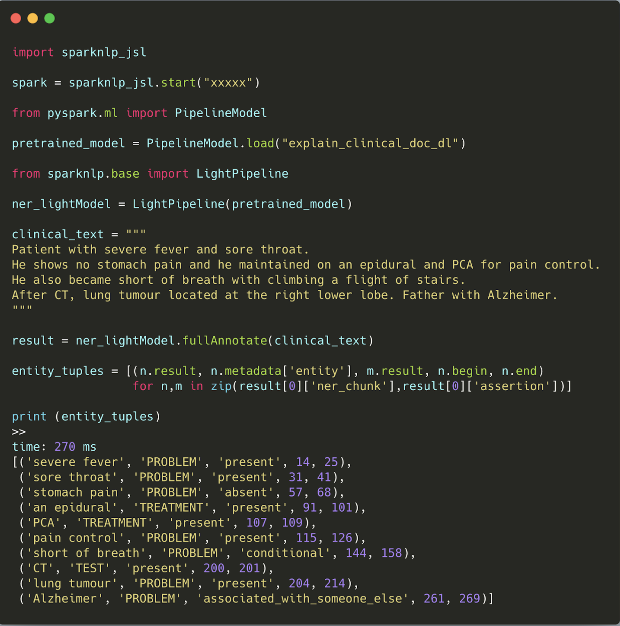
Pipeline descriptions:
explain_clinical_doc_carp
a pipeline with ner_clinical, assertion_dl, re_clinical and ner_posology. It will extract clinical and medication entities, assign assertion status and find relationships between clinical entities.
explain_clinical_doc_era
a pipeline with ner_clinical_events, assertion_dl and re_temporal_events_clinical. It will extract clinical entities, assign assertion status and find temporal relationships between clinical entities.
recognize_entities_posology
a pipeline with ner_posology. It will only extract medication entities.
More information and examples are available in this Python Colab Notebook.
… And More
We now have an out-of-the-box model for Named Entity Disambiguation.
Disambiguation models map words of interest, such as names of persons, locations and companies, from an input text document to corresponding unique entities in a target Knowledge Base (KB).

Also, due to ongoing requests about Clinical Entity Resolvers, we released a notebook to let you see how to train an entity resolver using an open-source dataset based on SNOMED-CT:
Last but not least: Spark NLP for Healthcare 2.6 is based on Spark NLP 2.6 – which means that all the new functionality of the latest open-source library is integrated & tested here, including:
- New multi-label document classifier
- New unsupervised keyword extraction
- BERT sentence embeddings
- 28 new pre-trained transformers: Small BERT, Covid BERT, ELECTRA
- 110 new pre-trained models
Enjoy our NLP solutions for healthcare! We’re already hard at work on the new release and welcome your feedback.
Our commitment to advancing Generative AI in Healthcare means continually enhancing tools like a Healthcare Chatbot, allowing for more precise information retrieval and improved patient engagement through cutting-edge NLP solutions.




























