Accurate Extraction of Response to Treatment Indications in Oncology
Accurate assessment of patient response to cancer treatment is paramount in guiding clinical decision-making and optimizing therapeutic outcomes. While large language models (LLMs) have demonstrated impressive capabilities in various natural language processing (NLP) tasks, their performance in this domain has been limited by the inherent complexities of medical language and the nuances involved in interpreting clinical narratives.
The ability to precisely comprehend the intricate details documented in clinical reports is essential for informing subsequent treatment decisions, adjusting therapeutic strategies, and ultimately improving patient outcomes. John Snow Labs provides models renowned for their exceptional accuracy in information extraction and classifying patient responses from unstructured texts, marking a significant stride forward in the application of NLP to cancer care.
This post covers using pretrained models in Healthcare NLP library from John Snow Labs to evaluate a patient’s response to cancer treatment. We will discuss identifying keywords or phrases NER in text data that are crucial in understanding the progress/changes in patient’s cancer treatment history and additionally, evaluate the response to the applied treatment by using a text classification model.
Let us start with a short Spark NLP introduction and then discuss the details of response to cancer treatment with some solid results.
Spark NLP & LLM
The Healthcare Library is a powerful component of John Snow Labs’ Spark NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,200 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Spark NLP and related tools.
John Snow Labs also offers periodic certification trainings to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
One crucial application area where the John Snow Labs Healthcare Library can play a pivotal role is in assessing patient response to cancer treatment. Accurately evaluating how an individual responds to therapy is paramount in guiding clinical decision-making and optimizing therapeutic outcomes. However, extracting this information from unstructured clinical documents can be challenging due to the complexities of medical language and nuances involved in interpreting such texts.
There are multiple approaches for evaluating a patient’s response to cancer treatment using NLP. The first approach involves using NER models to extract relevant oncology-related concepts and entities from unstructured clinical text. This allows systems to pinpoint key information pertaining to the patient’s condition, treatment regimen, and outcomes.
The second approach applies a text classification model to analyze the overall content and context of the clinical narratives. By training these models on labelled datasets, they can assess the effectiveness and efficiency of the prescribed cancer treatment based on the language used to describe the patient’s progress, symptoms, and other relevant factors.
In this blog post, we’ll explore the potential of John Snow Labs’ NER and text classification models as precise solutions for addressing this crucial concern.
Extracting Oncology-Related Entities from Clinical Notes
In this part, we’ll delve into how the John Snow Labs Healthcare NLP Library extracts oncology-related entities (NERs) from clinical notes.
By harnessing the power of more than 200 oncology-specific models, healthcare professionals can efficiently identify and extract vital information pertaining to cancer diagnosis, treatment, and patient outcomes from complex clinical narratives.
Spark NLP uses pipelines and for extracting valuable information, only 6 stages will be needed. I utilized the ner_oncology model to extract entities related to oncology.
# Step 1: Transforms raw texts to `document`
document = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
# Step 2: Sentence Detection/Splitting
sentencer = SentenceDetectorDLModel.pretrained("sentence_detector_dl_healthcare","en","clinical/models")\
.setInputCols(["document"])\
.setOutputCol("sentence")
# Step 3: Tokenization
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")\
.setSplitChars(["-", "\/"])
# Step 4: Clinical Embeddings
embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical","en","clinical/models")\
.setInputCols(["sentence","token"])\
.setOutputCol("embeddings")
# Oncology Model
model = MedicalNerModel.pretrained("ner_oncology","en","clinical/models")\
.setInputCols(["sentence","token","embeddings"])\
.setOutputCol("ner_oncology")\
converter = NerConverterInternal()\
.setInputCols(["sentence","token","ner_oncology"])\
.setOutputCol("ner_oncology_chunk")
# Define the pipeline
pipeline = Pipeline(stages=[document, tokenizer, embeddings, model, converter])
# Create an empty dataframe
empty_df = spark.createDataFrame([['']]).toDF("text")
# Fit the dataframe to the pipeline to get the model
pipelineModel = pipeline.fit(empty_df)
Let’s utilize this clinical text sample to assess the effectiveness of the model.
sample_text = """A 65-year-old woman underwent a computed tomography (CT) scan of the abdomen and pelvis, which showed a complex ovarian mass. A Pap smear performed one month later was positive for atypical glandular cells suspicious for adenocarcinoma. The pathologic specimen showed extension of the tumor throughout the fallopian tubes, appendix, omentum, and 5 out of 5 enlarged lymph nodes. The final pathologic diagnosis of the tumor was stage IIIC papillary serous ovarian adenocarcinoma. Two months later, the patient was diagnosed with lung metastases. """
Let’s use LightPipeline here to extract the entities. LightPipeline is a Spark NLP specific Pipeline class equivalent to the Spark ML Pipeline. The difference is that its execution does not hold to Spark principles, instead it computes everything locally (but in parallel) in order to achieve fast results when dealing with small amounts of data.
light_model = LightPipeline(pipelineModel)
light_result_onc = light_model.fullAnnotate(sample_text)
chunks = []
entities = []
sentence= []
begin = []
end = []
confidence = []
for n in light_result_onc[0]['ner_oncology_chunk']:
begin.append(n.begin)
end.append(n.end)
chunks.append(n.result)
entities.append(n.metadata['entity'])
sentence.append(n.metadata['sentence'])
confidence.append(n.metadata["confidence"])
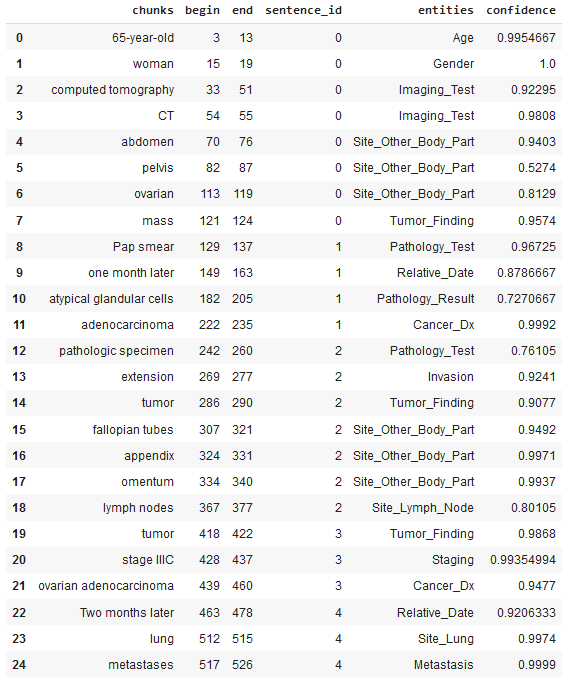
df_oncology = pd.DataFrame({'chunks':chunks, 'begin': begin, 'end':end,
'sentence_id':sentence, 'entities':entities, 'confidence':confidence})
df_oncology.head()
After applying the ner_oncology model, the following relevant medical concepts were automatically identified and extracted from the sample clinical notes:
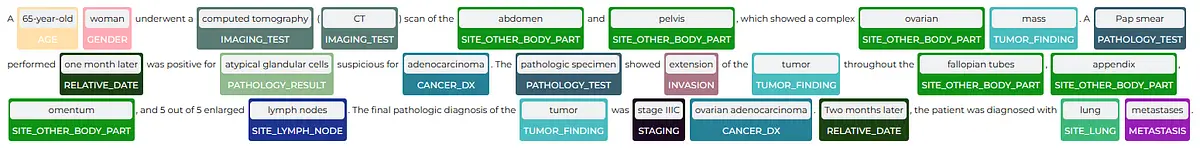
The ability to quickly visualize the entities generated using Spark NLP is a very useful feature for speeding up the development process as well as for understanding the obtained results. Spark NLP Display is an open-source python library for visualizing the extracted and labelled entities generated with Spark NLP. The NerVisualizer annotator highlights the extracted named entities and also displays their labels as decorations on top of the analyzed text.
from sparknlp_display import NerVisualizer visualiser = NerVisualizer() visualiser.display(light_result_onc[0], label_col='ner_oncology_chunk', document_col='document')
By highlighting these entities, we gain deeper insights into the patient’s condition, treatment plan, and overall prognosis, empowering healthcare professionals with valuable information to guide personalized care and optimize therapeutic outcomes in oncology practice.
Assessing Patient Response to Cancer Treatment from Clinical Reports
While NER models can extract the oncology concepts from clinical notes, understanding treatment response often requires analyzing the full context of the narrative. Text classification models can be employed to assess whether the overall content of a patient’s file indicates a positive response, or disease progression. By training on annotated datasets where clinical experts have labeled files based on treatment outcomes, these models learn predictive patterns from the language used to describe status, symptoms, imaging findings and other key factors. When applied to new unlabeled files, the models can categorize them at high accuracy. This automated classification can help triage cases for further review and streamline cohort identification for research. However, achieving high accuracy requires high-quality training data and careful handling of real-world language nuances in this domain.
To evaluate the performance of text classification model for this critical task, John Snow Labs data scientists trained and tested several models on a curated dataset of over 5,000 patient files. Each file was manually labeled by Medical Doctors as belonging to one of two categories based on treatment response — ‘The patient responded to treatment’, or ‘The patient did not respond to treatment’. The top performing model was a BERT-based architecture fine-tuned on this specialized dataset.
Let us use the Oncological Response to Treatment Classifier to get accurate label assignments for the pieces of clinical text shown below:
sample_texts = [
["Contrast-enhanced MRI of the brain showed no change in the size of the glioblastoma, suggesting stable disease post-temozolomide therapy."],
["The breast ultrasound after neoadjuvant chemotherapy displayed a decrease in the primary lesion size from 3 cm to 1 cm, suggesting a favorable response to treatment. The skin infection is also well controlled with multi-antibiotic approach. "],
["MRI of the pelvis indicated no further progression of endometriosis after laparoscopic excision and six months of hormonal suppression therapy."],
["A repeat endoscopy revealed healing gastric ulcers with new signs of malignancy or H. pylori infection. Will discuss the PPI continuum."],
["Dynamic contrast-enhanced MRI of the liver revealed no significant reduction in the size and number of hepatic metastases following six months of targeted therapy with sorafenib."],
["Digital subtraction angiography of the cerebral vessels displayed further aneurysmal dilation and new vascular abnormalities after endovascular coiling of a cerebral aneurysm, indicating a unsuccessful intervention."],
["The patient's repeat spirometry tests demonstrated non-significant improvement in both FEV1 and FVC, suggesting ineffective control of asthma symptoms with even maximally optimized inhaler therapy. Continuum will discuss."]
]
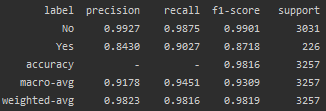
The accuracy of this model in predicting the response to the treatment applied is evident from the benchmarking results provided:
For text classification by using the Spark NLP annotator MedicalBertForSequenceClassification, the pipeline is even shorter:
# Step 1: Transforms raw texts to `document`
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
# Stage 2: Tokenization
tokenizer = Tokenizer()\
.setInputCols(["document"])\
.setOutputCol("token")
# Stage 3:Text Classification
sequenceClassifier = MedicalBertForSequenceClassification.pretrained("bert_sequence_classifier_response_to_treatment", "en", "clinical/models")\
.setInputCols(["document","token"])\
.setOutputCol("prediction")
pipeline = Pipeline(
stages=[
document_assembler,
tokenizer,
sequenceClassifier
])
# Generate Spark Dataframe from Sample Texts
sample_data = spark.createDataFrame(sample_texts).toDF("text")
# Fit the dataframe to the pipeline and get predictions
result = pipeline.fit(sample_data).transform(sample_data)
result.select("text", "prediction.result", 'prediction.metadata').show(truncate = 100)
After applying the text classification model, the sample clinical texts were categorized with confidence values as indicating either a response to treatment or showing no response.

The model demonstrated strong ability to distinguish between responding and non-responding cases based on the clinical notes. These results show the feasibility of using tailored text classification models as an initial triaging step before more comprehensive human review. With an accuracy over 93%, the model could automatically surface high-risk non-responding cases for prioritized evaluation by the oncology team. Continued iterative training as more labeled data becomes available could further boost performance.
Conclusion
In the critical domain of cancer care, where accurate assessment of response to treatment can impact treatment strategies and outcomes, John Snow Labs NER and Text Classification models achieved remarkable accuracy in extracting information and classifying patient response from complex clinical reports.
Using LLMs for evaluating a patient’s response to cancer treatment can enhance accuracy and provide contextual understanding. In general, larger models with more parameters tend to achieve better performance on many NLP tasks, but at a higher computational cost.
In conclusion, both approaches offer valuable insights into evaluating a patient’s response to cancer treatment. Utilizing NER and text classification models provides an efficient and accurate method for analyzing clinical text, offering insights into treatment outcomes. On the other hand, leveraging LLMs with billions of parameters promises enhanced accuracy due to their deeper understanding of context and semantics. However, this comes with an increase in cost, including computational resources and infrastructure requirements. Ultimately, the choice between these approaches depends on the specific needs of the healthcare setting, balancing the trade-offs between accuracy and cost-effectiveness.





























