Discover how Pretrained Zero-Shot Named Entity Recognition (NER) models simplify the recognition of entities in multiple domains with minimal configuration. Recent advancements in pretrained zero-shot NER models and their ease in being applied across different datasets and applications are described in the article.
Introduction to Pretrained Zero-Shot NER for Healthcare NLP
Named Entity Recognition (NER) is a cornerstone of NLP tasks, yet traditional NER approaches often require extensive domain-specific training to achieve high performance. The Pretrained Zero-Shot NER models remove this barrier by providing pretrained, fine-tuned language models that can adapt to custom entity definitions without additional training.
Besides the diverse Named Entity Recognition (NER) models available in our Models Hub, like the Healthcare NLP MedicalNerModel using Bidirectional LSTM-CNN architecture and BertForTokenClassification, our library also includes powerful rule-based annotators such as ContextualParser, TextMatcher, RegexMatcher, and EntityRulerInternal.
Now, John Snow Labs is newly announcing 10 additional Pretrained Zero-Shot NER models, further expanding the capabilities of the strong models mentioned above. These models offer exceptional adaptability in identifying entities across diverse texts. Here’s what they offer:
- Leverage robust contextual understanding from pretrained language models.
- Support user-defined labels, extending their capabilities beyond the default entity types.
- Allow for rapid adaptation to new domains and datasets.
This blog post will demonstrate the capabilities of these models and guide you through their implementation in Healthcare NLP.
What is Pretrained Zero-Shot Named Entity Recognition?
Pretrained Zero-Shot NER is a versatile approach to entity recognition that eliminates the need for fine-tuning on domain-specific data. These models:
- Identify entities based on predefined labels.
- Provide flexibility to define any entity type relevant to your use case.
Key Features:
- Pretrained Flexibility: Models are fine-tuned on in-house annotations but can adapt to new entity labels defined by users.
- Custom Labels: Users can define entity types and hypotheses, making the models suitable for various use cases.
- Rapid Adaptation: Ideal for scenarios requiring fast setup and deployment.
For more details, examples, and usage, please check this Colab notebook on Zero-Shot Clinical NER.

10 New Pretrained ZeroShotNER Named Entity Recognition Models that are Already Finetuned on In-house Annotations
Implementation in Healthcare NLP
Let’s walk through a practical implementation of ZeroShotNerModel using a sample notebook.
Step 1: Set Up the Environment
First, we need to set up the Spark NLP Healthcare library. Follow the detailed instructions provided in the official documentation.
Additionally, refer to the Healthcare NLP GitHub repository for sample notebooks demonstrating setup on Google Colab under the “Colab Setup” section.
# Install the johnsnowlabs library to access Spark-OCR and Spark-NLP for Healthcare, Finance, and Legal.
! pip install -q johnsnowlabs
from google.colab import files
print('Please Upload your John Snow Labs License using the button below')
license_keys = files.upload()
from johnsnowlabs import nlp, medical
# After uploading your license run this to install all licensed Python Wheels and pre-download Jars the Spark Session JVM
nlp.settings.enforce_versions=True
nlp.install(refresh_install=True)
from johnsnowlabs import nlp, medical
import pandas as pd
# Automatically load license data and start a session with all jars user has access to
spark = nlp.start()
Step 2: Define Your Text and Pipeline
We’ll create a pipeline using the zeroshot_ner_deid_subentity_merged_medium model:
document_assembler = nlp.DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentence_detector = nlp.SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = nlp.Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
labels = ['DOCTOR', 'PATIENT', 'AGE', 'DATE', 'HOSPITAL', 'CITY',
'STREET', 'STATE', 'COUNTRY', 'PHONE', 'IDNUM', 'EMAIL',
'ZIP', 'ORGANIZATION', 'PROFESSION', 'USERNAME']
pretrained_zero_shot_ner = medical.PretrainedZeroShotNER()\
.pretrained("zeroshot_ner_deid_subentity_merged_medium", "en", "clinical/models")\
.setInputCols("sentence", "token")\
.setOutputCol("entities")\
.setPredictionThreshold(0.5)\
.setLabels(labels)
ner_converter = medical.NerConverterInternal()\
.setInputCols("sentence", "token", "entities")\
.setOutputCol("ner_chunk")
pipeline = nlp.Pipeline().setStages([
document_assembler,
sentence_detector,
tokenizer,
pretrained_zero_shot_ner,
ner_converter
])
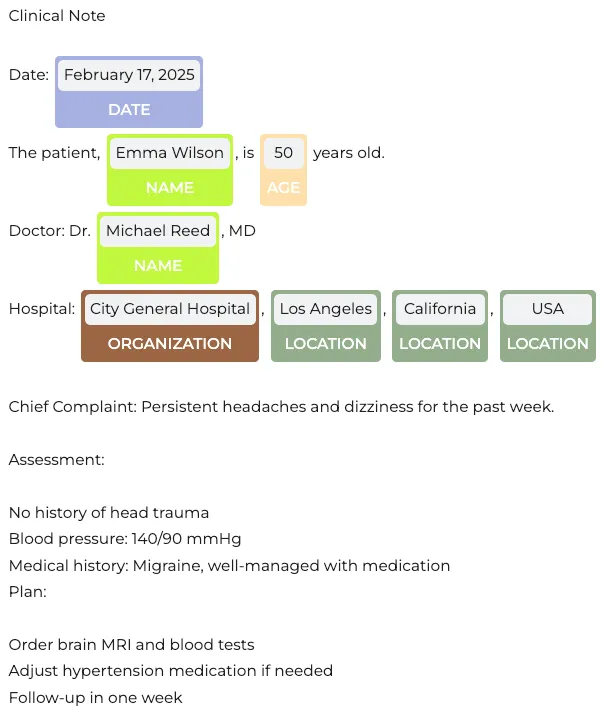
We’ll use sample medical text to demonstrate entity recognition:
text = """Clinical Note
Date: February 17, 2025
The patient, Emma Wilson, is 50 years old.
Doctor: Dr. Michael Reed, MD
Hospital: City General Hospital, Los Angeles, California, USA
Chief Complaint: Persistent headaches and dizziness for the past week.
Assessment:
No history of head trauma
Blood pressure: 140/90 mmHg
Medical history: Migraine, well-managed with medication
Plan:
Order brain MRI and blood tests
Adjust hypertension medication if needed
Follow-up in one week
"""
data = spark.createDataFrame([[text]]).toDF("text")
result = pipeline.fit(data).transform(data)
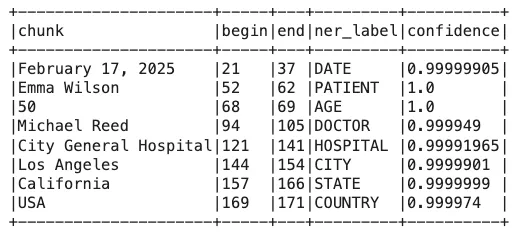
Step 3: Extract Entities
Extract entities from the text and view results:
result = pipeline.fit(data).transform(data)
result.selectExpr("explode(ner_chunk) as ner_chunk")\
.selectExpr("ner_chunk.result as chunk",
"ner_chunk.begin",
"ner_chunk.end",
"ner_chunk.metadata['entity'] as ner_label",
"ner_chunk.metadata['confidence'] as confidence").show(100, truncate=False)
One of the standout features of the Zero-Shot NER models is their flexibility. Users are not restricted to predefined labels; you have the freedom to define and use labels tailored to your specific use case. By providing your desired labels, the model will seamlessly adapt to predict them.
You can customize the labels in your pipeline to better align with your needs. For example, similar entities can be grouped together, such as DOCTOR → NAME and PATIENT → NAME, which we will apply in the pipeline below. Likewise, location-related entities like CITY, STREET, STATE, and COUNTRY can be grouped under LOCATION for a more streamlined classification.
Here’s how you can customize labels in your pipeline:
labels = ['NAME', 'AGE', 'DATE', 'LOCATION', 'IDNUM',
'ORGANIZATION', 'PROFESSION']
pretrained_zero_shot_ner = medical.PretrainedZeroShotNER()\
.pretrained("zeroshot_ner_deid_subentity_merged_medium", "en", "clinical/models")\
.setInputCols("sentence", "token")\
.setOutputCol("ner")\
.setPredictionThreshold(0.5)\
.setLabels(labels)
ner_converter = medical.NerConverterInternal()\
.setInputCols("sentence", "token", "ner")\
.setOutputCol("ner_chunk")
pipeline = nlp.Pipeline().setStages([
document_assembler,
sentence_detector,
tokenizer,
pretrained_zero_shot_ner,
ner_converter
])
result = pipeline.fit(data).transform(data)
result.selectExpr("explode(ner_chunk) as ner_chunk")\
.selectExpr("ner_chunk.result as chunk",
"ner_chunk.begin",
"ner_chunk.end",
"ner_chunk.metadata['entity'] as ner_label").show(100, truncate=False)
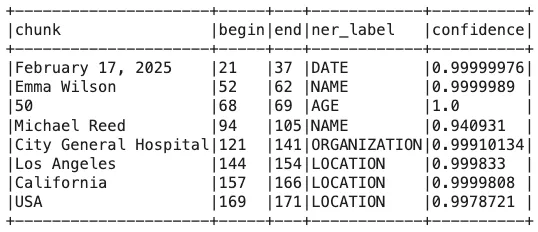
Customized Label Results
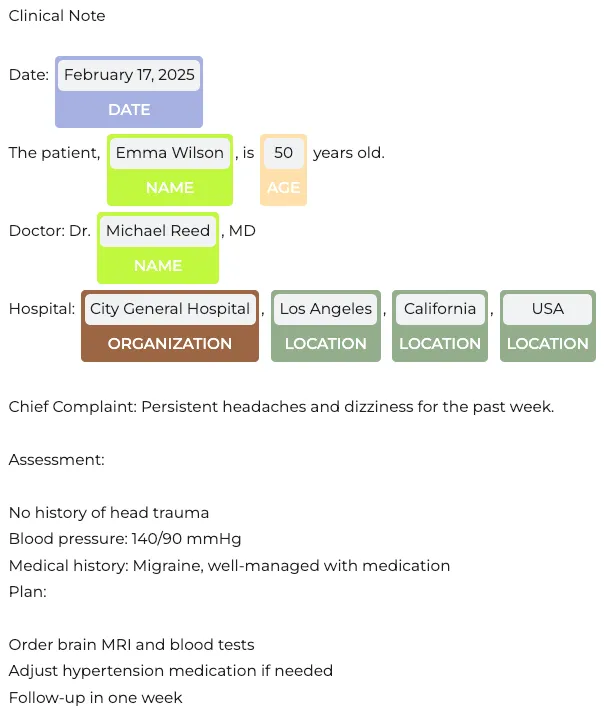
Visualization of Customized Label Results
When customizing entity labels in a pipeline, it’s essential to compare the modified results with the original annotations. For instance, in the initial pipeline, Emma Wilson was identified as PATIENT, but after customization, she is now labeled as NAME. Similarly, Michael Reed, previously classified as DOCTOR, is now recognized under NAME.
The changes extend to organizations and locations as well. City General Hospital, which was originally tagged as HOSPITAL, is now categorized as ORGANIZATION. Likewise, Los Angeles (previously labeled as CITY), California (STATE), and USA (COUNTRY) are now grouped under a broader LOCATION category.
These modifications highlight the flexibility of entity recognition models, allowing tailored outputs based on specific project needs.
#Replaced Entities DOCTOR, PATIENT => NAME, HOSPITAL => ORGANIZATION, CITY, STREET, STATE, COUNTRY=> LOCATION
Conclusion
The Pretrained Zero-Shot Named Entity Recognition (NER) models represent a significant leap forward in NLP technology, especially in domains like healthcare where accuracy and adaptability are paramount. By eliminating the need for extensive domain-specific training, these models make it easier than ever to identify and classify entities across a variety of datasets and industries.
With pre-defined labels from high-end NER models or with custom labels for domain-specific applications, companies can achieve flexibility like never before. Such flexibility is incredibly helpful in quickly adapting to specific datasets and overcoming varied requirements without ever compromising on accuracy.
Furthermore, the integration of zero-shot models with Healthcare NLP pipelines gives practitioners access to state-of-the-art NLP enhancements without losing valuable setup time or resource investment. From managing highly complex medical writing to moving into entirely new domains, these models offer unparalleled productivity and performance.