Automated De-Identification, Consistent Obfuscation, and Regulatory Grade Validation of 2 Billion Patient Notes
Authors: Lindsay Mico (Providence Health), Mustafa Aytug Kaya (George Mason University), Nadaa Taiyab (Providence Health), David Talby (John Snow Labs), Tae Surh (Providence Health), Yuqing Guo (Providence Health), Vivek Tomer (Providence Health), Robert Kramer (Providence Health).
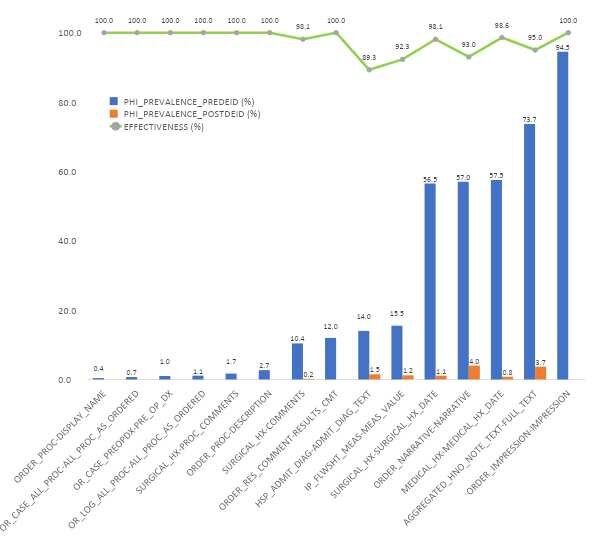
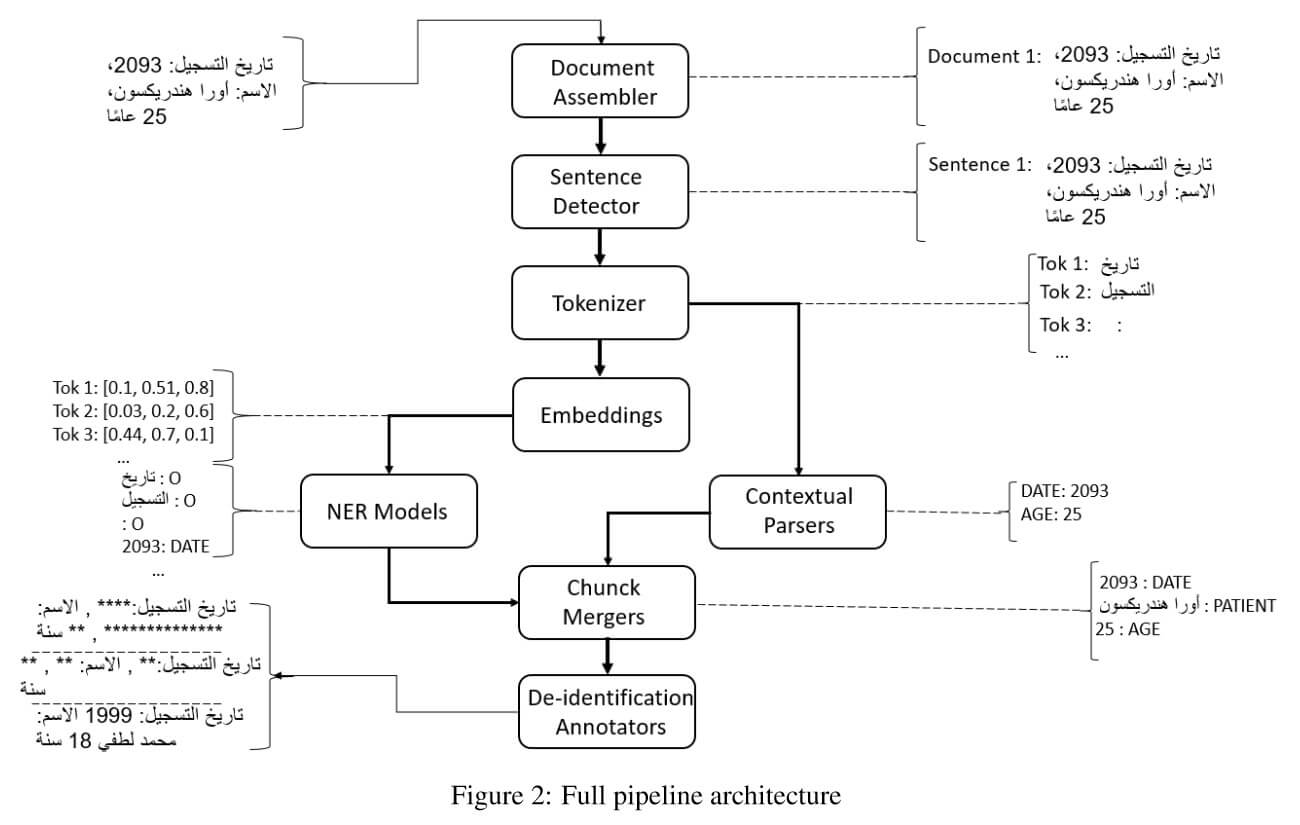
Rich Large, diverse collections of anonymous patient data—including text, numbers, and images— are essential to advancing a broad range of causes, from clinical decision support and real-world evidence to population health and hospital operations. This study presents a novel system used to automatically de-identify unstructured clinical text from 2 billion patient notes, using consistent obfuscation and tokenization to link them into a unified longitudinal dataset. To the best of our knowledge, this is the first such system to be externally certified for regulatory-grade accuracy on real-world data at this scale. The system is based on proprietary medical language models and the modified Spark NLP – a distributed computing NLP framework for efficient execution on large clusters of commodity hardware. It satisfies the Expert Determination de-identification criteria under HIPAA ( Health Insurance Portability and Accountability Act) Privacy Rules, establishing a baseline requirement of 5% PHI prevalence both in aggregate and per record. It achieves 99% Protected Health Information (PHI) obfuscation, and achieves 100% masking or shifting of target data fields. This level of accuracy surpasses even that of a triple manual review by 3 human annotators.
Exploring NER Potential and the Value of Tailored NLP Pipelines for Radiology, Pathology, and Progress Notes in Clinical Decision Support: Quantitative Study
Authors: Veysel Kocaman (John Snow Labs), Fu-Yuan Cheng (Mount Sinai Hospital), Julio Bonis (John Snow Labs), Ganesh Raut (New Jersey Institute of Technology).
JMIR AI 4 (2025)
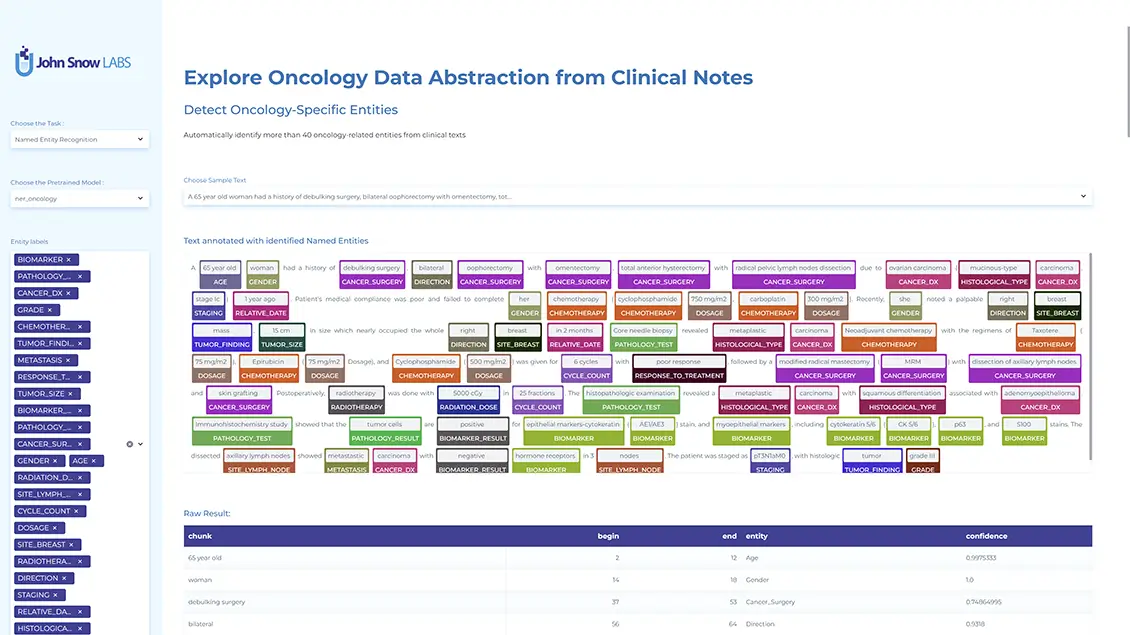
Clinical notes contain valuable but unstructured data, posing challenges for real-time analysis due to jargon, abbreviations, and synonyms. We examined the data curation, technology, and workflow of a named entity recognition (NER) pipeline within a clinical decision support tool. Notes from 5,000 patients (progress, radiology, pathology) were analyzed in five batches. We measured runtime, CPU, memory, and evaluated NER precision and assertion model performance against expert annotations. Using Spark NLP pretrained clinical models on 138,250 notes, we achieved high NER precision, peaking at 0.989 for procedures, and assertion model accuracy of 0.889. Entity density varied widely: progress notes contained 4× more entities per sentence than radiology notes and 16x more than pathology notes. These findings highlight long-tail distributions in note characteristics and the need for specialized NER models tailored to different note types to improve performance across diverse clinical scenarios.
CLEVER: Clinical Large Language Model Evaluationby Expert Review
Authors: Veysel Kocaman, Mustafa Kaya, Andrei Ferrer, David Talby.
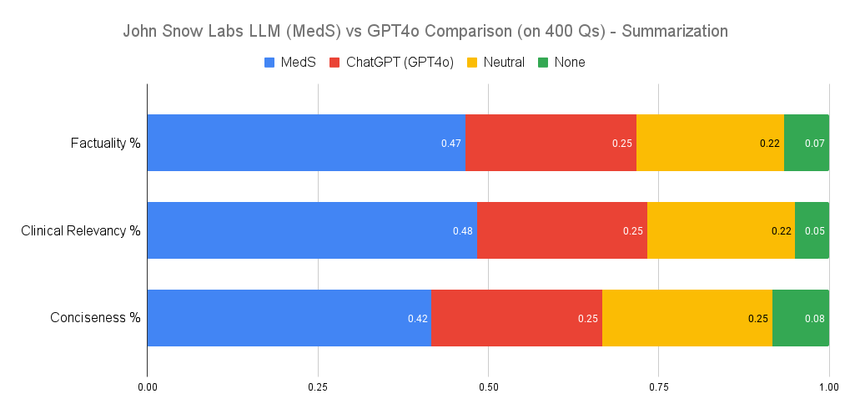
The proliferation of both general-purpose and healthcare-specific Large Language Models (LLMs) has intensified the challenge of rigorous benchmarking. Existing evaluation methods face critical limitations: data contamination undermines the validity of public benchmarks, self-preference biases LLM-as-a-judge approaches, and current tasks do not fully reflect real-world clinical applications. To address these issues, we introduce CLEVER, a blind, randomized, preference-based evaluation methodology conducted by practicing medical doctors on task-specific assessments. We apply CLEVER to compare GPT-4o with two healthcare-specific LLMs (8B and 70B parameters) across three tasks: medical summarization, clinical information extraction, and biomedical question answering. Results reveal that domain-specific small LLMs outperform GPT-4o by 45% to 92% in factuality, clinical relevance, and conciseness, while maintaining comparable performance in open-ended medical Q&A. These findings challenge the assumption that larger general-purpose models inherently excel in specialized domains.
An Obfuscation Algorithm Designed to Support Machine Learning with Real World Evidence
Authors: John Philip,Ilya Sher, Rohan Singh, Sribatsa Das, Mrunmayi Deshpande.
Clinical Research Informatics.
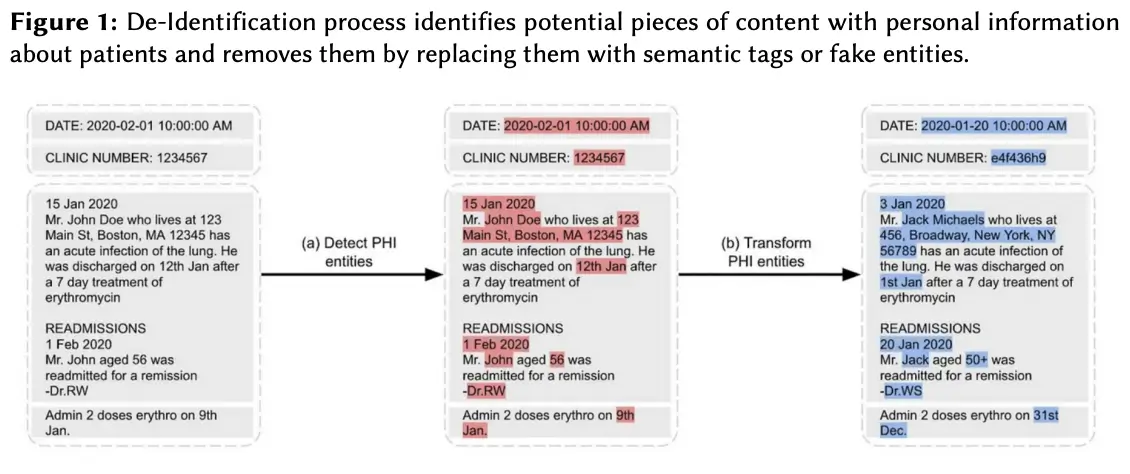
To support HIPAA-compliant machine learning with real-world clinical data, Memorial Sloan Kettering developed a patient-centric obfuscation algorithm that deidentifies unstructured electronic health records. Using John Snow Labs’ NLP tools and expert annotation, PHI labels were validated across ~14,000 documents. The algorithm preserves document structure, consistency, and typographical quirks while ensuring high-quality obfuscation.
Can Zero-Shot Commercial APIs Deliver Regulatory-Grade Clinical Text DeIdentification?
Authors: Veysel Kocaman, Muhammed Santas, Yigit Gul, Mehmet Butgul, David Talby.
Accepted at Text2Story Workshop at ECIR 2025.
We evaluate the performance of four leading solutions for de-identification of unstructured medical text – Azure Health Data Services, AWS Comprehend Medical, OpenAI GPT-4o, and John Snow Labs – on a ground truth dataset of 48 clinical documents annotated by medical experts. The analysis, conducted at both entity-level and token-level, suggests that John Snow Labs’ Medical Language Models solution achieves the highest accuracy, with a 96% F1-score in protected health information (PHI) detection, outperforming Azure (91%), AWS (83%), and GPT-4o (79%).
John Snow Labs is not only the only solution which achieves regulatory-grade accuracy (surpassing that of human experts) but is also the most cost-effective solution: It is over 80% cheaper compared to Azure and GPT-4o, and is the only solution not priced by token. Its fixed-cost local deployment model avoids the escalating per-request fees of cloud-based services, making it a scalable and economical choice.
The Importance of Information Extraction from Unstructured Clinical Data in Pharmacoepidemiology
Authors: Veysel Kocaman, Muhammed Santas, Yigit Gul, Mehmet Butgul, David Talby.
Accepted to: the 2025 PHUSE/FDA Computational Science Symposium (CSS).
This study we will discuss the use of natural language processing in a pharmacoepidemiology study, done as part of the FDA Sentinel Innovation Center MOSAIC-NLP (Multi-source Observational Safety study for Advanced Information Classification using NLP). Electronic health record (EHR) data linked to claims were used to examine the association of montelukast with neuropsychiatric events in patients with asthma. The study analyzed data from ~17 million notes from a cohort of 109,076 patients from 112 health systems in Oracle Real World Data. Information extracted from unstructured clinical notes used medical language models from John Snow Labs to enhance outcome identification and confounder control.
The study found that neuropsychiatric events, including agitation, anxious feelings, confusion, irritability, memory problems, and self-harm may be undercounted using only structured data from EHR and claims, as the number of observed suicidality/self-harm events doubled with the addition of unstructured EHR data. Further, events such as irritability, agitation, and memory problems were only detected in unstructured data. This study illustrates the importance of unstructured data especially related to mental health outcomes.

Beyond Negation Detection: Comprehensive Assertion Detection Models for Clinical NLP.
Authors: Veysel Kocaman, David Talby, Yigit Gul, M. Aytug Kaya, Hasham Ul Haq, Mehmet Butgul, Cabir Celik.
Accepted at Text2Story Workshop at ECIR 2025.
Assertion status detection is a critical yet often overlooked component of clinical NLP, essential for accurately attributing extracted medical facts. Past studies have narrowly focused on negation detection, leading to underperforming commercial solutions such as AWS Medical Comprehend, Azure AI Text Analytics, and GPT-4o due to their limited domain adaptation. To address this gap, we developed state-of-the-art assertion detection models, including fine-tuned LLMs, transformer-based classifiers, few-shot classifiers, and deep learning (DL) approaches. We evaluated these models against cloud-based commercial API solutions, the legacy rule-based NegEx approach, and GPT-4o. Our fine-tuned LLM achieves the highest overall accuracy (0.962), outperforming GPT-4o (0.901) and commercial APIs by a notable margin, particularly excelling in Present (+4.2%), Absent (+8.4%), and Hypothetical (+23.4%) assertions.
Our DL-based models surpass commercial solutions in Conditional (+5.3%) and Associated-with-Someone-Else (+10.1%) categories, while the few-shot classifier offers a lightweight yet highly competitive alternative (0.929), making it ideal for resource-constrained environments. Integrated within Spark NLP, our models consistently outperform black-box commercial solutions while enabling scalable inference and seamless integration with medical NER, Relation Extraction, and Terminology Resolution. These results reinforce the importance of domain-adapted, transparent, and customizable clinical NLP solutions over general-purpose LLMs and proprietary APIs
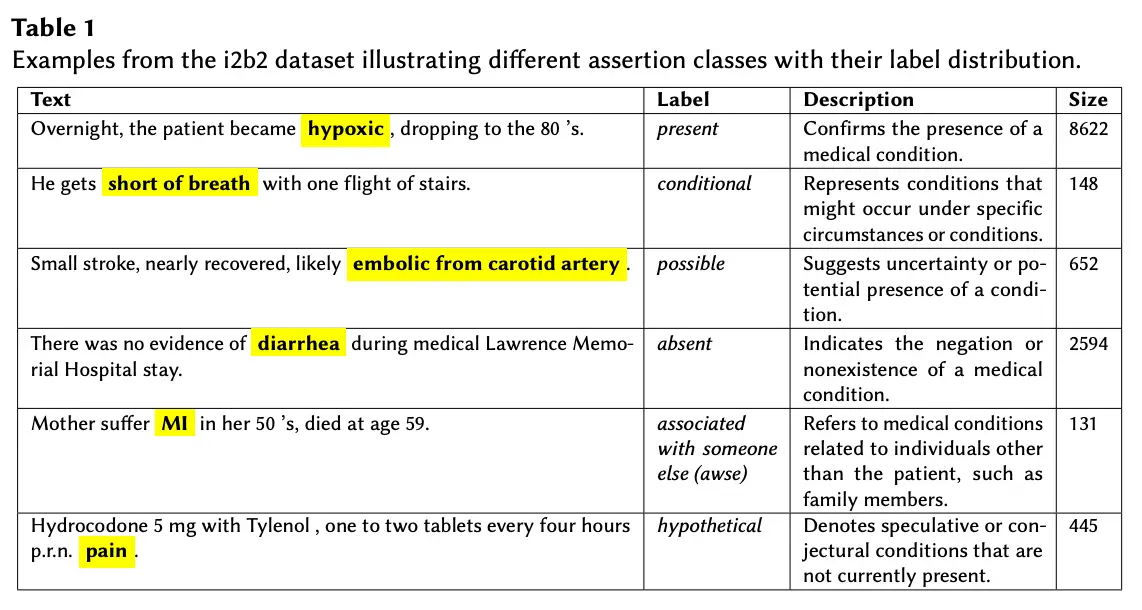
Holistic Evaluation of Large Language Models: Assessing Robustness, Accuracy, and Toxicity for Real-World Applications.
Authors: David Cecchini, Arshaan Nazir, Kalyan Chakravarthy, Veysel Kocaman.
Proceedings of the 4th Workshop on Trustworthy Natural Language Processing (TrustNLP 2024).
Large Language Models (LLMs) have been widely used in real-world applications. However, as LLMs evolve and new datasets are released, it becomes crucial to build processes to evaluate and control the models’ performance. In this paper, we describe how to add Robustness, Accuracy, and Toxicity scores to model comparison tables, or leaderboards. We discuss the evaluation metrics, the approaches considered, and present the results of the first evaluation round for model Robustness, Accuracy, and Toxicity scores.
Our results show that GPT 4 achieves top performance on robustness and accuracy test, while Llama 2 achieves top performance on the toxicity test. We note that newer open-source models such as open chat 3.5 and neural chat 7B can perform well on these three test categories. Finally, domain-specific tests and models are also planned to be added to the leaderboard to allow for a more detailed evaluation of models in specific areas such as healthcare, legal, and finance.
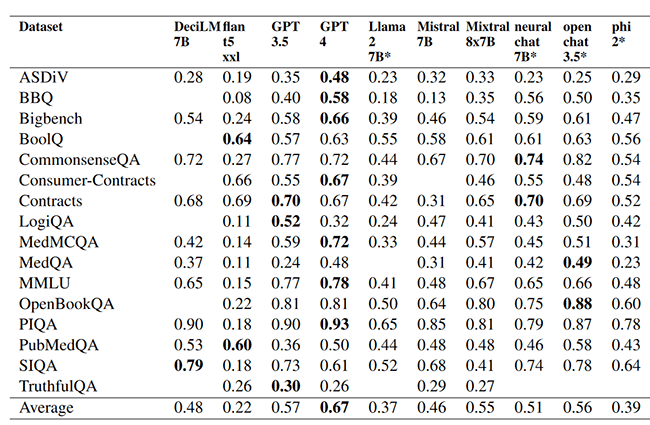
Chemotimeline chronicles – leveraging large language models for temporal relations extraction in oncological electronic health records.
Authors: Vishakha Sharma, Andres Fernandez, Andrei Ioanovici, David Talby, Frederik Buijs.
Proceedings of the 6th Clinical Natural Language Processing Workshop, 2024.
Automatic generation of chemotherapy treatment timelines from electronic health records (EHRs) notes not only streamlines clinical workflows but also promotes better coordination and improvements in cancer treatment and quality of care. This paper describes the submission to the Chemotimelines 2024 shared task that aims to automatically build a chemotherapy treatment timeline for each patient using their complete set of EHR notes, spanning various sources such as primary care provider, oncology, discharge summaries, emergency department, pathology, radiology, and more. We report results from two large language models (LLMs), namely Llama 2 and Mistral 7B, applied to the shared task data using zero-shot prompting.
Summary of the National Cancer Institute 2023 Virtual Workshop on Medical Image De-identification
Authors: David Clunie, Fred Prior, Michael Rutherford, Jiri Dobes et al.
Journal of Imaging Informatics in Medicine, 2024.
De-identification of medical images intended for research is a core requirement for data-sharing initiatives, particularly as the demand for data for artificial intelligence (AI) applications grows. The Center for Biomedical Informatics and Information Technology (CBIIT) of the US National Cancer Institute (NCI) convened a virtual workshop with the intent of summarizing the state of the art in de-identification technology and processes and exploring interesting aspects of the subject.
This paper summarizes the highlights of the first day of the workshop, the recordings, and presentations of which are publicly available for review. The topics covered included the report of the Medical Image De-Identification Initiative (MIDI) Task Group on best practices and recommendations, tools for conventional approaches to de-identification, international approaches to de-identification, and an industry panel.
Efficient schema-less text-to-SQL conversion using large language models
Authors: Youssef Mellah, Veysel Kocaman, Hasham Ul Haq, David Talby
Artificial Intelligent in Health, 2024.
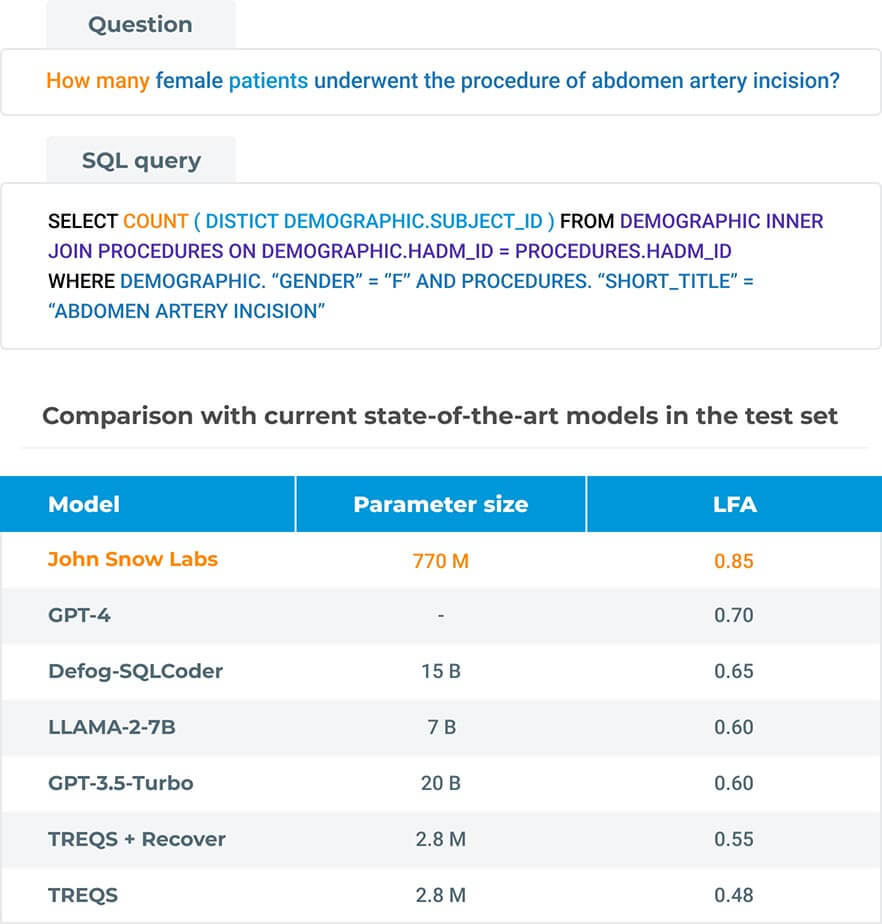
Large language models (LLMs) are increasingly being applied to several tasks including text-to-SQL (the process of converting natural language to SQL queries). While most studies revolve around training LLMs on large SQL corpora for better generalization and then perform prompt engineering during inference, we investigate the notion of training LLMs for schema-less prompting. In particular, our approach uses simple natural language questions as input without any additional knowledge about the database schema. By doing so, we demonstrate that smaller models paired with simpler prompts result in considerable performance improvement while generating SQL queries.
Our model, based on the Flan-T5 architecture, achieves logical form accuracy (LFA) of 0.85 on the MIMICSQL dataset, significantly outperforming current state-of-the-art models such as Defog-SQL-Coder, GPT-3.5-Turbo, LLaMA-2-7B and GPT-4. This approach reduces the model size, lessening the amount of data and infrastructure cost required for training and serving, and improves the performance to enable the generation of much complex SQL queries.
LangTest: A Comprehensive Evaluation Library for Custom LLM and NLP Models
Authors: Arshaan Nazir, Thadaka Kalyan Chakravarthy, David Amore Cecchini, Rakshit Khajuria, Prikshit Sharma, Ali Tarik Mirik, Veysel Kocaman, David Talby
Introduction of LangTest, an open-source Python toolkit for evaluating LLMs and NLP models 2024.
The use of natural language processing (NLP) models, including the more recent large language models (LLM) in real-world applications obtained relevant success in the past years. To measure the performance of these systems, traditional performance metrics such as accuracy, precision, recall, and f1-score are used. Although it is important to measure the performance of the models in those terms, natural language often requires an holistic evaluation that consider other important aspects such as robustness, bias, accuracy, toxicity, fairness, safety, efficiency, clinical relevance, security, representation, disinformation, political orientation, sensitivity, factuality, legal concerns, and vulnerabilities.
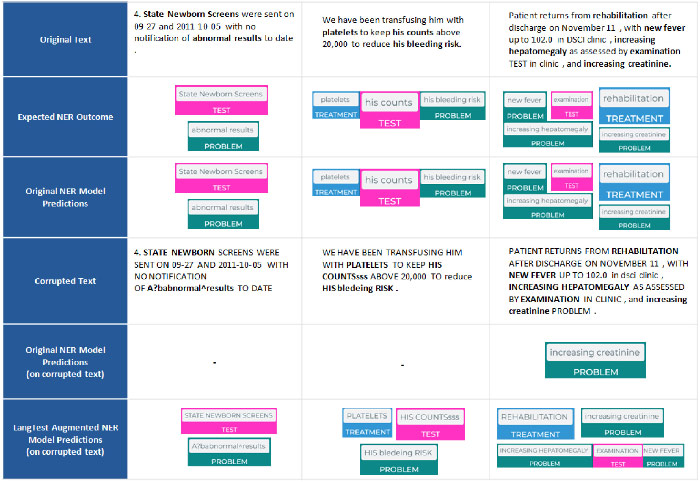
To address the gap, we introduce LangTest, an open source Python toolkit, aimed at reshaping the evaluation of LLMs and NLP models in real-world applications. The project aims to empower data scientists, enabling them to meet high standards in the ever-evolving landscape of AI model development. Specifically, it provides a comprehensive suite of more than 60 test types, ensuring a more comprehensive understanding of a model’s behavior and responsible AI use. In this experiment, a Named Entity Recognition (NER) clinical model showed significant improvement in its capabilities to identify clinical entities in text after applying data augmentation for robustness.
Beyond Accuracy: Automated De-Identification of Large Real-World Clinical Text Datasets
Authors: Veysel Kocaman, Hasham Ul Haq, David Talby
Machine Learning for Health (ML4H) 2023.
Also presented as a poster at ISPOR Europe 2023.
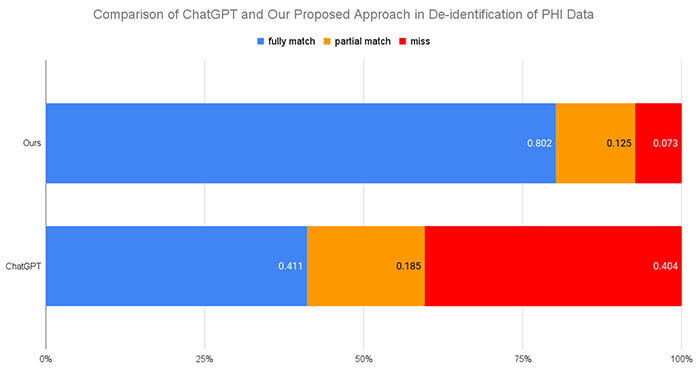
Recent research advances achieve human-level accuracy for de-identifying free-text clinical notes on research datasets, but gaps remain in reproducing this in large real-world settings. This paper summarizes lessons learned from building a system used to e-identify over one billion real clinical notes, in a fully automated way, that was independently certified by multiple organizations for production use.
A fully automated solution requires a very high level of accuracy that does not require manual review. A hybrid context-based model architecture is described, which outperforms a Named Entity Recogniton (NER)-only model by 10% on the i2b2-2014 benchmark. The proposed system makes 50%, 475%, and 575% fewer errors than the comparable AWS, Azure, and GCP services respectively while also outperforming ChatGPT by 33%. It exceeds 98% coverage of sensitive data across 7 European languages, without a need for fine tuning.
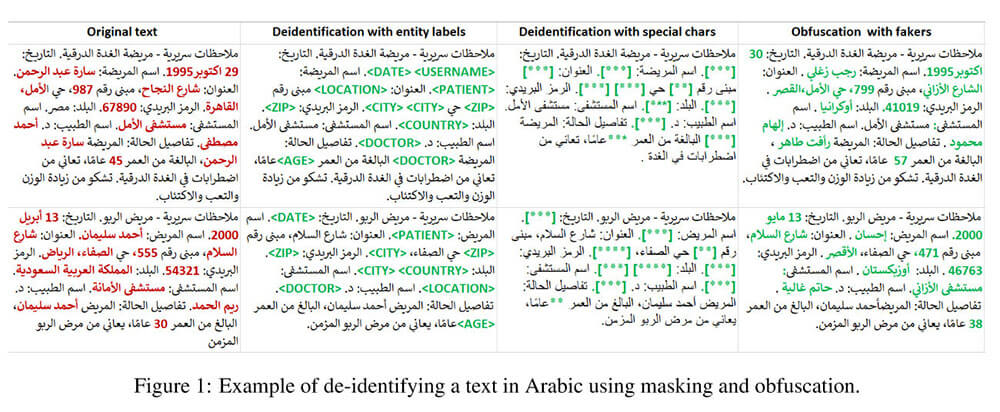
Automated De-Identification of Arabic Medical Records
Authors: Veysel Kocaman, Youssef Mellah, Hasham Haq, and David Talby
Association for Computational Linguistics 2023.
Proceedings of ArabicNLP 2023.
As Electronic Health Records (EHR) become ubiquitous in healthcare systems worldwide, including in Arabic-speaking countries, the dual imperative of safeguarding patient privacy and leveraging data for research and quality improvement grows. This paper presents a first-of-its-kind automated de-identification pipeline for medical text specifically tailored for the Arabic language.
This includes accurate medical Named Entity Recognition (NER) for identifying personal information; data obfuscation models to replace sensitive entities with fake entities; and an implementation that natively scales to large datasets on commodity clusters. This research makes two contributions. First, we adapt two existing NER architectures— BERT For Token Classification (BFTC) and BiLSTM-CNN-Char – to accommodate the unique syntactic and morphological characteristics of the Arabic language. Comparative analysis suggests that BFTC models outperform Bi-LSTM models, achieving higher F1 scores for both identifying and redacting personally identifiable information (PII) from Arabic medical texts.
Second, we augment the deep learning models with a contextual parser engine to handle commonly missed entities. Experiments show that the combined pipeline demonstrates superior performance with micro F1 scores ranging from 0.94 to 0.98 on the test dataset, which is a translated version of the i2b2 2014 de-identification challenge, across 17 sensitive entities. This level of accuracy is in line with that achieved with manual de-identification by domain experts, suggesting that a fully automated and scalable process is now viable.
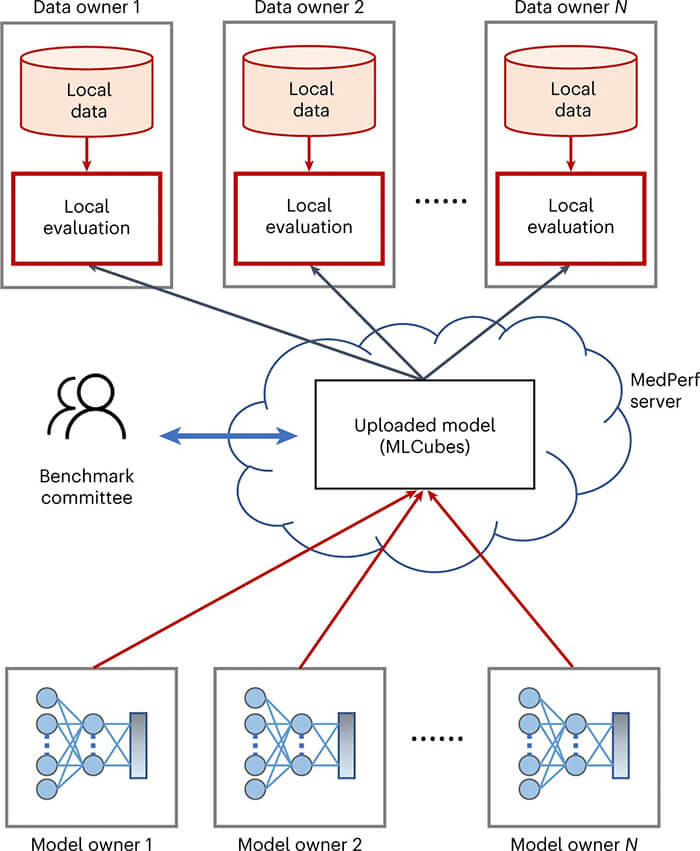
Federated Benchmarking of Medical Artificial Intelligence with MedPerf
Authors: Alexandros Karargyris, Renato Umeton, Micah J. Sheller
Nature Machine Intelligence 2023.
Medical artificial intelligence (AI) has tremendous potential to advance healthcare by supporting and contributing to the evidence-based practice of medicine, personalizing patient treatment, reducing costs, and improving both healthcare provider and patient experience. Unlocking this potential requires systematic, quantitative evaluation of the performance of medical AI models on large-scale, heterogeneous data capturing diverse patient populations. Here, to meet this need, we introduce MedPerf, an open platform for benchmarking AI models in the medical domain. MedPerf focuses on enabling federated evaluation of AI models, by securely distributing them to different facilities, such as healthcare organizations.
This process of bringing the model to the data empowers each facility to assess and verify the performance of AI models in an efficient and human-supervised process, while prioritizing privacy. We describe the current challenges healthcare and AI communities face, the need for an open platform, the design philosophy of MedPerf, its current implementation status and real-world deployment, our roadmap and, importantly, the use of MedPerf with multiple international institutions within cloud-based technology and on-premises scenarios. Finally, we welcome new contributions by researchers and organizations to further strengthen MedPerf as an open benchmarking platform.
Advanced Queries, Search and Biomedical Predictions Leveraging Disparate Oncology Unstructured Data Compendia
Authors: V. Sharma, A. Thomas, V. Vettrivel, David Talby, A. Vladimirova
Annals of Oncology 2022.
In healthcare product development and research, teams invest huge amounts of time to study publications and other relevant sources. There is a need for novel solutions to efficiently and reliably extract information from multiple clinical data sources, in addition to generating new insights which can only be achieved through structuring textual information and accessible intelligent synthesis across multiple relevant data sources. We are developing a cloud-based solution where data from heterogeneous sources is structured, integrated and harmonized, and users can easily leverage the combined database to answer domain-specific questions and generate insights efficiently.
Knowledge graphs provide us the advantage to leverage relationships in addition to concepts in the context of heterogeneous data. We leveraged graph and NLP (Natural Language Processing) methods to build a domain-specific knowledge graph. We extracted the biomedically-relevant subset of Wikidata and augmented it from the biomedical literature (PubMed), clinical trials (clinicaltrials.gov) and NIH grants.

Detecting Adverse Drug events in Dermatology Through Natural Language Processing of Physician Notes
Authors: Vikas Kumar, Lawrence Rasouliyan, Veysel Kocaman, and David Talby
ICPE 2022, the 38th International Conference on Pharmacoepidemiology and Therapeutic Risk Management (ICPE).
Pharmacoepidemiology studies often have missing data that are nonmonotone (i.e., data missing in an arbitrary pattern). It is well known that complete case analysis (CC) reduces precision and may result in bias. Previously, inverse probability weighting (IPW) for nonmonotone missingness could not be easily implemented in standard software, but a new approach for estimating the missingness weights (unconstrained maximum likelihood estimator, UMLE) overcomes this difficulty.
To compare performance of IPW by UMLE and multiple imputation (MI) to address nonmonotone missingness when estimating the average treatment effect.

Deeper Clinical Document Understanding Using Relation Extraction
Authors: Hasham Ul Haw, Veysel Kocaman and David Talby
Accepted to SDU (Scientific Document Understanding) workshop at AAAI 2022.
The surging amount of biomedical literature & digital clinical records presents a growing need for text mining techniques that can not only identify but also semantically relate entities in unstructured data. In this paper we propose a text mining framework comprising of Named Entity Recognition (NER) and Relation Extraction (RE) models, which expands on previous work in three main ways. First, we introduce two new RE model architectures — an accuracy-optimized one based on BioBERT and a speed-optimized one utilizing crafted features over a Fully Connected Neural Network (FCNN).
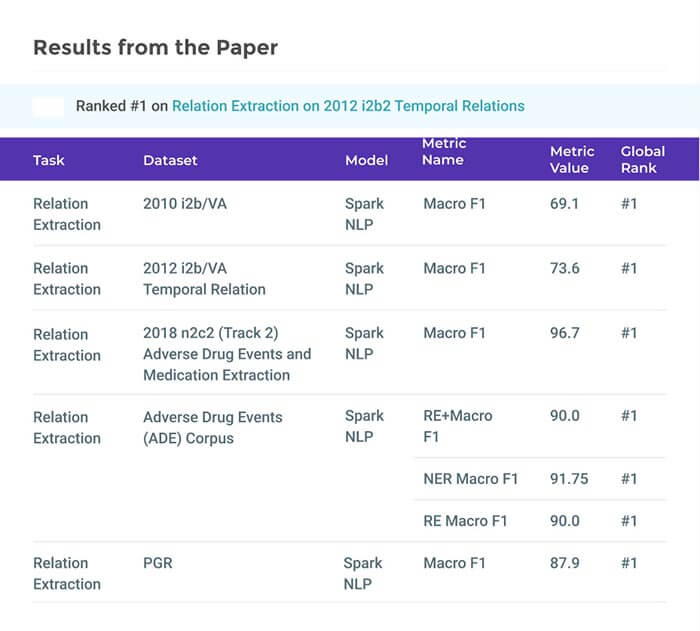
Second, we evaluate both models on public benchmark datasets and obtain new state-of-the-art F1 scores on the 2012 i2b2 Clinical Temporal Relations challenge (F1 of 73.6, +1.2% over the previous SOTA), the 2010 i2b2 Clinical Relations challenge (F1 of 69.1, +1.2%), the 2019 Phenotype-Gene Relations dataset (F1 of 87.9, +8.5%), the 2012 Adverse Drug Events Drug-Reaction dataset (F1 of 90.0, +6.3%), and the 2018 n2c2 Posology Relations dataset (F1 of 96.7, +0.6%). Third, we show two practical applications of this framework — for building a biomedical knowledge graph and for improving the accuracy of mapping entities to clinical codes. The system is built using the Spark NLP library which provides a production-grade, natively scalable, hardware-optimized, trainable & tunable NLP framework.
Mining Adverse Drug Reactions from Unstructured Mediums at Scale
Authors: Hasham Ul Haw, Veysel Kocaman and David Talby
Accepted to the 6th International Workshop on Health Intelligence at AAAI 2022.
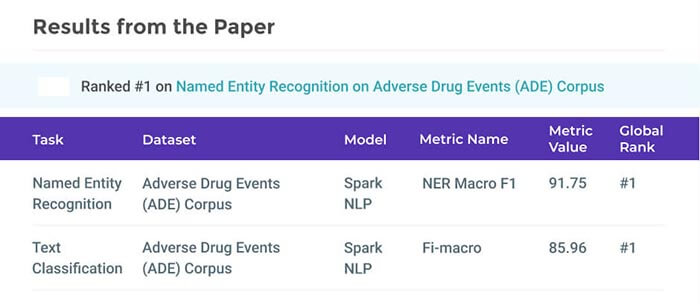
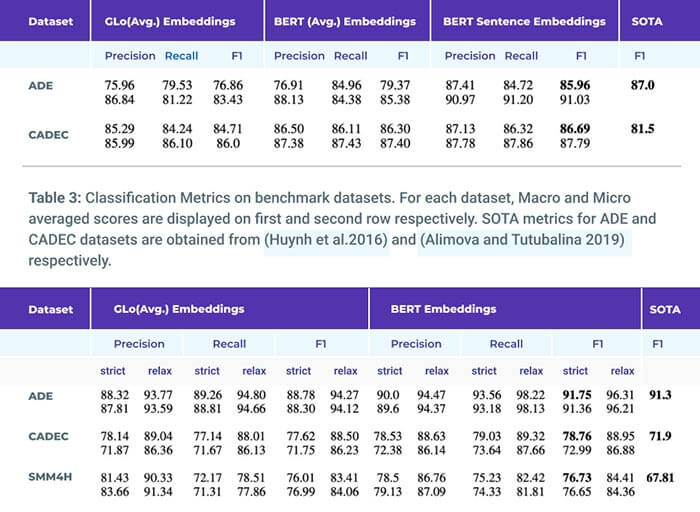
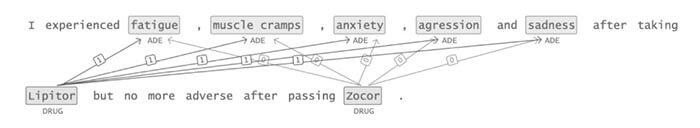
Adverse drug reactions / events (ADR/ADE) have a major impact on patient health and health care costs. Detecting ADR’s as early as possible and sharing them with regulators, pharma companies, and healthcare providers can prevent morbidity and save many lives. While most ADR’s are not reported via formal channels, they are often documented in a variety of unstructured conversations such as social media posts by patients, customer support call transcripts, or CRM notes of meetings between healthcare providers and pharma sales reps. In this paper, we propose a natural language processing (NLP) solution that detects ADR’s in such unstructured free-text conversations, which improves on previous work in three ways. First, a new Named Entity Recognition (NER) model obtains new state-of-the-art accuracy for ADR and Drug entity extraction on the ADE, CADEC, and SMM4H benchmark datasets (91.75%, 78.76%, and 83.41% F1 scores respectively).
Second, two new Relation Extraction (RE) models are introduced – one based on BioBERT while the other utilizing crafted features over a Fully Connected Neural Network (FCNN) – are shown to perform on par with existing state-of-the-art models, and outperform them when trained with a supplementary clinician-annotated RE dataset. Third, a new text classification model, for deciding if a conversation includes an ADR, obtains new state-of-the-art accuracy on the CADEC dataset (86.69% F1 score). The complete solution is implemented as a unified NLP pipeline in a production-grade library built on top of Apache Spark, making it natively scalable and able to process millions of batch or streaming records on commodity clusters.
Biomedical Named Entity Recognition at Scale
Authors: Veysel Kocaman and David Talby
Accepted for presentation and inclusion in CADL 2020 (International Workshop on Computational Aspects of Deep Learning), in conjunction with ICPR 2020.
Named entity recognition (NER) is a widely applicable natural language processing task and building block of question answering, topic modeling, information retrieval, etc. In the medical domain, NER plays a crucial role by extracting meaningful chunks from clinical notes and reports, which are then fed to downstream tasks like assertion status detection, entity resolution, relation extraction, and de-identification. Reimplementing a Bi-LSTM-CNN-Char deep learning architecture on top of Apache Spark, we present a single trainable NER model that obtains new state-of-the-art results on seven public biomedical benchmarks without using heavy contextual embeddings like BERT.
This includes improving BC4CHEMD to 93.72% (4.1% gain), Species800 to 80.91% (4.6% gain), and JNLPBA to 81.29% (5.2% gain). In addition, this model is freely available within a production-grade code base as part of the open-source Spark NLP library; can scale up for training and inference in any Spark cluster; has GPU support and libraries for popular programming languages such as Python, R, Scala and Java; and can be extended to support other human languages with no code changes.
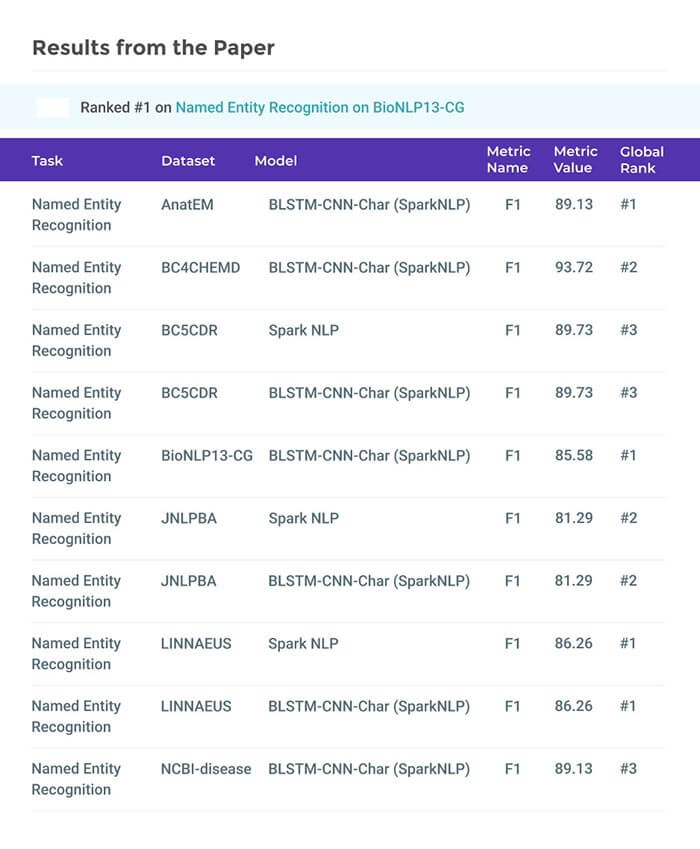
Clinical Application of Detecting COVID-19 Risks: A Natural Language Processing Approach
Authors: Syed Raza Bashir, Shaina Raza,Veysel Kocaman and Urooj Qamar
Abstract: The clinical application of detecting COVID-19 factors is a challenging task. The existing named entity recognition models are usually trained on a limited set of named entities. Besides clinical, the non-clinical factors, such as social determinant of health (SDoH), are also important to study the infectious disease. In this paper, we propose a generalizable machine learning approach that improves on previous efforts by recognizing a large number of clinical risk factors and SDoH.
The novelty of the proposed method lies in the subtle combination of a number of deep neural networks, including the BiLSTM-CNN-CRF method and a transformer-based embedding layer. Experimental results on a cohort of COVID-19 data prepared from PubMed articles show the superiority of the proposed approach. When compared to other methods, the proposed approach achieves a performance gain of about 1–5% in terms of macro- and micro-average F1 scores. Clinical practitioners and researchers can use this approach to obtain accurate information regarding clinical risks and SDoH factors, and use this pipeline as a tool to end the pandemic or to prepare for future pandemics.
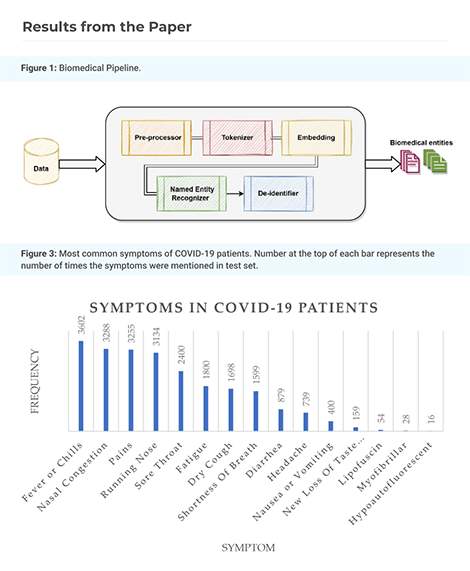
Improving Clinical Document Understanding on COVID-19 Research with Spark NLP
Authors: Veysel Kocaman and David Talby
Accepted to SDU (Scientific Document Understanding) workshop at AAAI 2021.
Following the global COVID-19 pandemic, the number of scientific papers studying the virus has grown massively, leading to increased interest in automated literate review. We present a clinical text mining system that improves on previous efforts in three ways. First, it can recognize over 100 different entity types including social determinants of health, anatomy, risk factors, and adverse events in addition to other commonly used clinical and biomedical entities.
Second, the text processing pipeline includes assertion status detection, to distinguish between clinical facts that are present, absent, conditional, or about someone other than the patient. Third, the deep learning models used are more accurate than previously available, leveraging an integrated pipeline of state-of-the-art pretrained named entity recognition models, and improving on the previous best performing benchmarks for assertion status detection.
We illustrate extracting trends and insights, e.g. most frequent disorders and symptoms, and most common vital signs and EKG findings, from the COVID-19 Open Research Dataset (CORD-19). The system is built using the Spark NLP library which natively supports scaling to use distributed clusters, leveraging GPUs, configurable and reusable NLP pipelines, healthcare specific embeddings, and the ability to train models to support new entity types or human languages with no code changes.
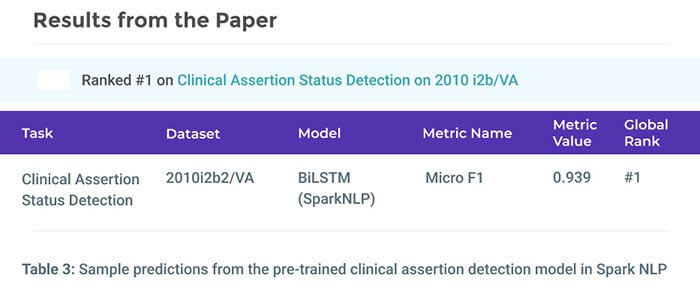
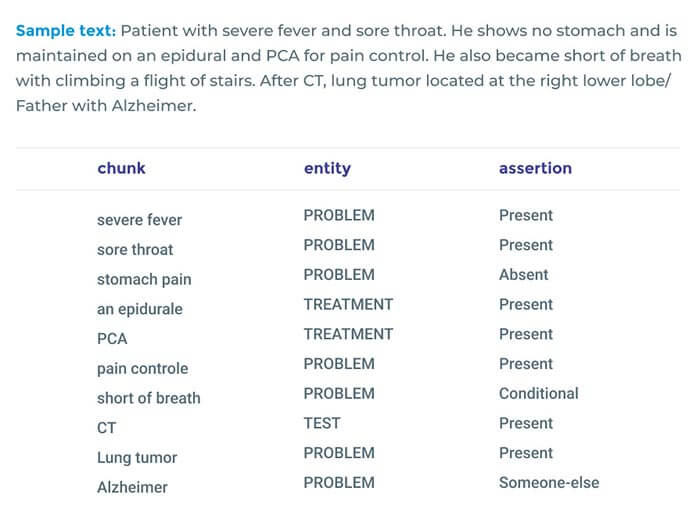
Accurate Clinical and Biomedical Named Entity Recognition at Scale
Authors: Veysel Kocaman and David Talby
Accepted to Software Impacts, July 2022.
Named entity recognition (NER) is one of the most important building blocks of NLP tasks in the medical domain by extracting meaningful chunks from clinical notes and reports, which are then fed to downstream tasks like assertion status detection, entity resolution, relation extraction, and de-identification. Due to the growing volume of healthcare data in unstructured format, an increasingly important challenge is providing high accuracy implementations of state-of-the-art deep learning (DL) algorithms at scale.
While recent advances in NLP like Transformers and BERT have pushed the boundaries for accuracy, these methods are significantly slow and difficult to scale on millions of records.
In this study, we introduce an agile, production-grade clinical and biomedical NER algorithm based on a modified BiLSTM-CNN-Char DL architecture built on top of Apache Spark.
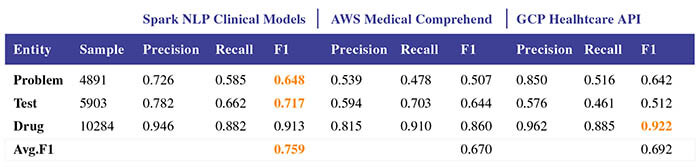
Our NER implementation establishes new state-of-the-art accuracy on 7 of 8 well-known biomedical NER benchmarks and 3 clinical concept extraction challenges: 2010 i2b2/VA clinical concept extraction, 2014 n2c2 de-identification, and 2018 n2c2 medication extraction. Moreover, clinical NER models trained using this implementation outperform the accuracy of commercial entity extraction solutions such AWS Medical Comprehend and Google Cloud Healthcare API by a large margin (8.9% and 6.7% respectively), without using memory-intensive language models.
The proposed model requires no handcrafted features or task-specific resources, requires minimal hyperparameter tuning for a given dataset from any domain, can be trained with any embeddings including BERT, and can be trained to support more human languages with no code changes. It is available within a production-grade code base as part of the Spark NLP library, the only open-source NLP library that can scale to make use of a Spark cluster for training and inference, has GPU support, and provides libraries for Python, R, Scala and Java.


Preparing for the next pandemic via transfer learning from existing diseases with hierarchical multi-modal BERT: a study on COVID-19 outcome prediction
Authors: Khushbu Agarwal, Sutanay Choudhury, Sindhu Tipirneni, Pritam Mukherjee, Colby Ham, Suzanne Tamang, Matthew Baker, Siyi Tang, Veysel Kocaman, Olivier Gevaert, Robert Rallo & Chandan K Reddy
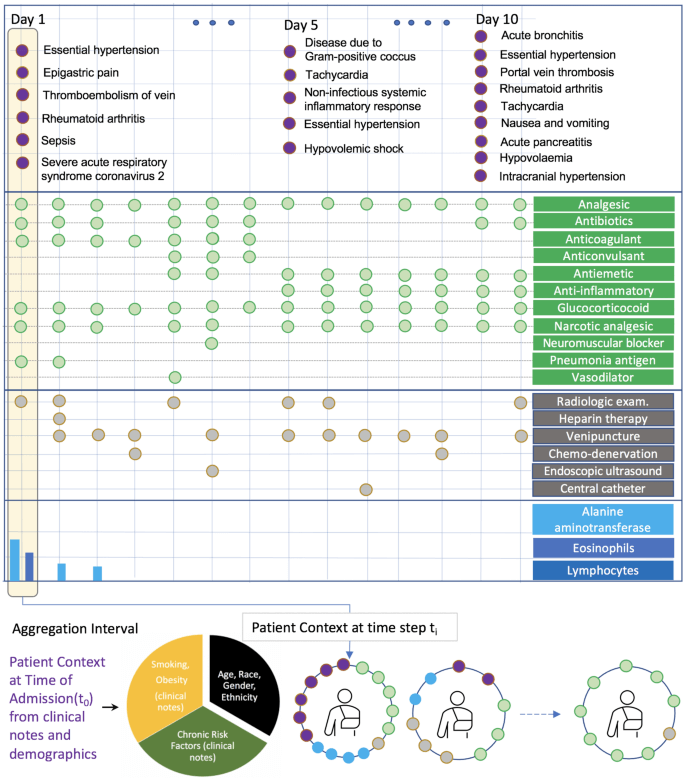
Developing prediction models for emerging infectious diseases from relatively small numbers of cases is a critical need for improving pandemic preparedness. Using COVID-19 as an exemplar, we propose a transfer learning methodology for developing predictive models from multi-modal electronic healthcare records by leveraging information from more prevalent diseases with shared clinical characteristics. Our novel hierarchical, multi-modal model (TRANSMED) integrates baseline risk factors from the natural language processing of clinical notes at admission, time-series measurements of biomarkers obtained from laboratory tests, and discrete diagnostic, procedure and drug codes.
We demonstrate the alignment of TRANSMED’s predictions with well-established clinical knowledge about COVID-19 through univariate and multivariate risk factor driven sub-cohort analysis. TRANSMED’s superior performance over state-of-the-art methods shows that leveraging patient data across modalities and transferring prior knowledge from similar disorders is critical for accurate prediction of patient outcomes, and this approach may serve as an important tool in the early response to future pandemics.
Spark NLP: A Versatile Solution for Structuring Data from Endoscopy Reports
Authors: Ioanovici, Andrei Constantin; Măruşteri, Ştefan Marius; Feier, Andrei Marian; Trambitas-Miron, Alina Dia.
Spark NLP: A Versatile Solution for Structuring Data from Endoscopy Reports
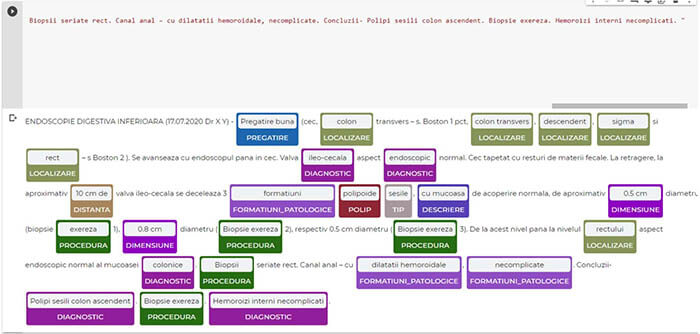
Artificial intelligence (AI) can be applied in the practice of gastroenterology to acquire and analyze information. Besides speed and duplicability, AI has the potential of also offering insight with results that surpass medical specialists. Natural language processing (NLP) is being used to extract information from text, organize and categorize documents. Processing unstructured data with NLP will result in structured data and medical codes can be extracted more easily (ICD10, medical procedure codes, etc) for reimbursement purposes among others. Recent research is studying the use of AI for automated interpretation of text from endoscopy and medical documents for better quality and patient phenotyping as well as enhanced detection and descriptions of endoscopic lesions such as colon polyps. In this paper, we present a method of extracting medical data using Spark NLP (John Snow Labs, DE, USA), by annotating endoscopy reports and training a model to automatically extract labels in order to obtain
structured medical data. This can be used in combination with other forms of structured data for an optimal and novel patient profiling.
Tracking the Evolution of COVID-19 via Temporal Comorbidity Analysis from Multi-Modal Data
Authors: Sutanay Choudhury, Khushbu Agarwal, Colby Ham, Pritam Mukherjee, Siyi Tang, Sindhu Tipirneni, Chandan Reddy, Suzanne Tamang, Robert Rallo, Veysel Kocaman
Presented at AMIA-2021 Annual Symposium.
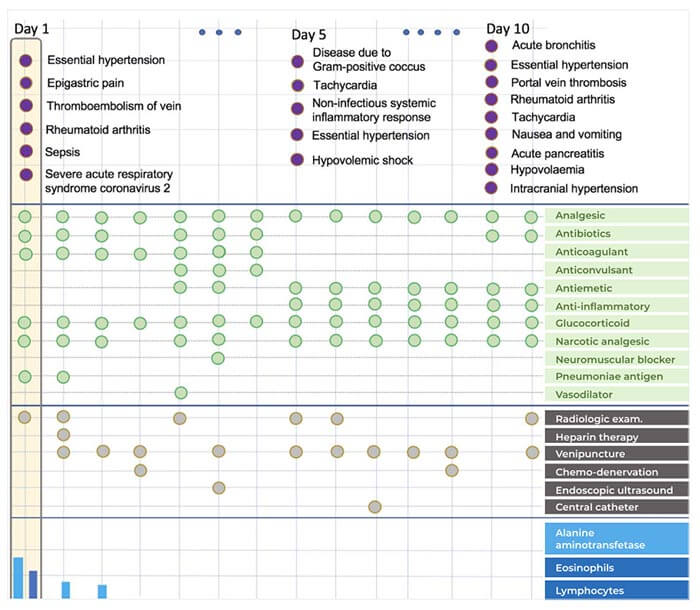
Developing prediction models for emerging infectious diseases from relatively small numbers of cases is a critical need for improving pandemic preparedness. Using COVID-19 as an exemplar, we propose a transfer learning methodology for developing predictive models from multi-modal electronic healthcare records by leveraging information from more prevalent diseases with shared clinical characteristics. Our novel hierarchical, multi-modal model (TRANSMED) integrates baseline risk factors from the natural language processing of clinical notes at admission, time-series measurements of biomarkers obtained from laboratory tests, and discrete diagnostic, procedure and drug codes.
We demonstrate the alignment of TRANSMED’s predictions with well-established clinical knowledge about COVID-19 through univariate and multivariate risk factor driven sub-cohort analysis. TRANSMED’s superior performance over state-of-the-art methods shows that leveraging patient data across modalities and transferring prior knowledge from similar disorders is critical for accurate prediction of patient outcomes, and this approach may serve as an important tool in the early response to future pandemics.
Connecting the dots in clinical document understanding with Relation Extraction at scale
Authors: Hasham Ul Haw, Veysel Kocaman and David Talby
Easy to use, scalable NLP framework that can leverage Spark. Introduction of BERT based Relation Extraction models. State-of-the-art performance on Named Entity Recognition and Relation Extraction. Reported SOTA performance of multiple public benchmark datasets. Application of these models on real-world use-cases.
We present a text mining framework based on top of the Spark NLP library — comprising of Named Entity Recognition (NER) and Relation Extraction (RE) models, which expands on previous work in three main ways. First, we release new RE model architectures that obtain state-of-the-art F1 scores on 5 out of 7 benchmark datasets. Second, we introduce a modular approach to train and stack multiple models in a single nlp pipeline in a production grade library with little coding. Third, we apply these models in practical applications including knowledge graph generation, prescription parsing, and robust ontology mapping.
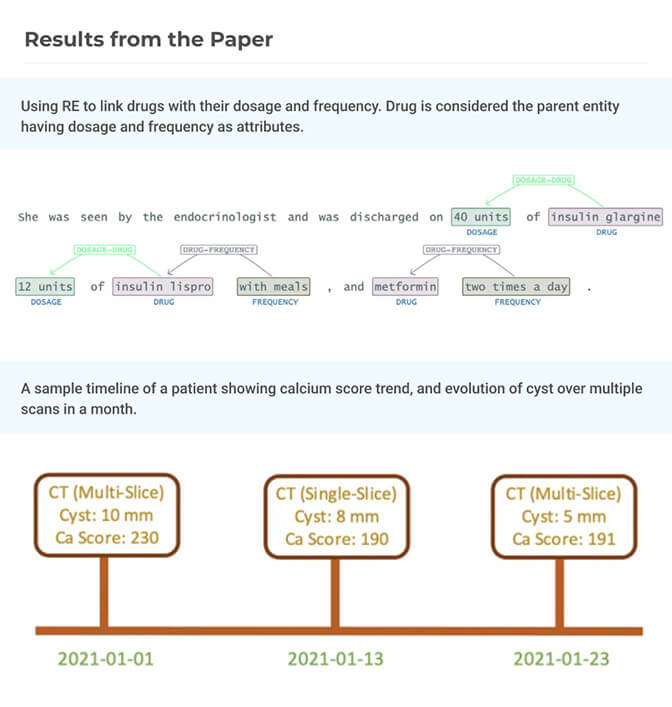
Spark NLP: Natural Language Understanding at Scale
Authors: Veysel Kocaman and David Talby
Accepted as a publication in Elsevier, Software Impacts Journal.
Spark NLP is a Natural Language Processing (NLP) library built on top of Apache Spark ML. It provides simple, performant and accurate NLP annotations for machine learning pipelines that can scale easily in a distributed environment. Spark NLP comes with 1100 pre trained pipelines and models in more than 192 languages. It supports nearly all the NLP tasks and modules that can be used seamlessly in a cluster. Downloaded more than 2.7 million times and experiencing nine times growth since January 2020, Spark NLP is used by 54% of healthcare organizations as the worlds most widely used NLP library in the enterprise.

Understanding COVID-19 News Coverage using Medical NLP
Authors: Ali Emre Varol, Veysel Kocaman, Hasham Ul Haq, David Talby
Proceedings of the Text2Story’22 Workshop, Stavanger (Norway).
Being a global pandemic, the COVID-19 outbreak received global media attention. In this study, we analyze news publications from CNN and The Guardian – two of the world’s most influential media organizations. The dataset includes more than 36,000 articles, analyzed using the clinical and biomedical Natural Language Processing (NLP) models from the Spark NLP for Healthcare library, which enables a deeper analysis of medical concepts than previously achieved. The analysis covers key entities and phrases, observed biases, and change over time in news coverage by correlating mined medical symptoms, procedures, drugs, and guidance with commonly mentioned demographic and occupational groups. Another analysis is of extracted Adverse Drug Events about drug and vaccine manufacturers, which when reported by major news outlets has an impact on vaccine hesitancy.
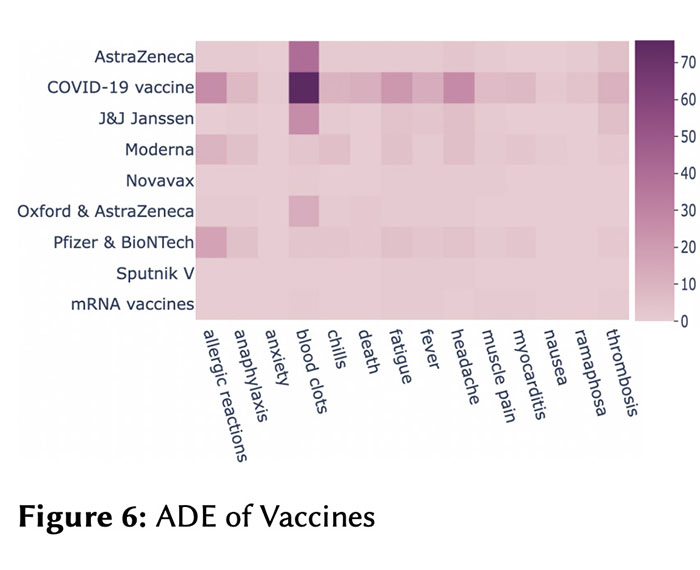
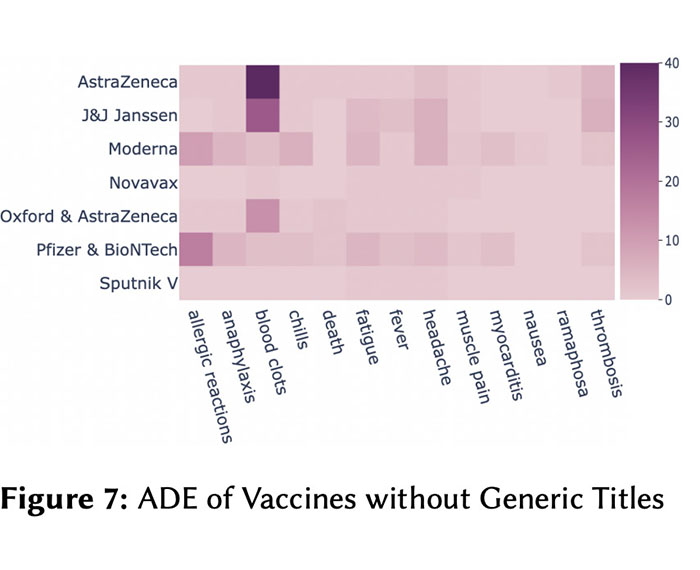
Biomedical Named Entity Recognition in Eight Languages with Zero Code Changes
Authors: Veysel Kocaman, Gursev Pirge, Bunyamin Polat, David Talby
Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2022), A Coruña, Spain, September 2022.
Named entity recognition (NER) is one of the most important building blocks of NLP tasks in the medical domain by extracting meaningful chunks from clinical notes and reports, which are then fed to downstream tasks like assertion status detection, entity resolution, relation extraction, and de-identification. Due to the growing volume of healthcare data in unstructured format, an increasingly important challenge is providing high accuracy implementations of state-of-the-art deep learning (DL) algorithms at scale. On the other hand, when it comes to low-resource languages, collecting high quality annotated data sets in the biomedical domain
is still a big challenge. In this study, we train production-grade biomedical NER models on eight different biomedical datasets published within the LivingNER competition [1].
Transformer based Bert for token classification and BiLSTM-CNN-Char based NER algorithms from Spark NLP library are utilized during this study and we trained 28 different NER models in total with decent accuracies (0.9243 F1 test score in Spanish) without changing the underlying DL architecture. The trained models are available within a production-grade code base as part of the Spark NLP library; can scale up for training and inference in any Spark cluster; has GPU support and libraries for popular programming languages such as Python, R, Scala and Java
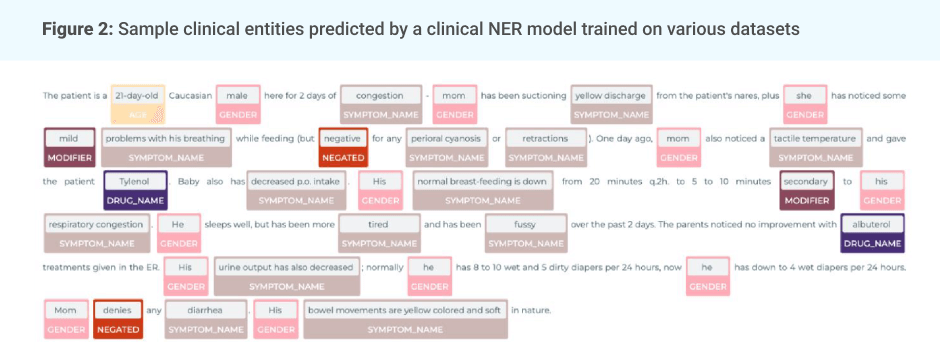























Social Media Mining for Health (#SMM4H) with Spark NLP.
Veysel Kocaman, Cabir Celik, Damla Gurbaz, Gursev Pirge, Bunyamin Polat, Halil Saglamlar, Meryem Vildan Sarikaya, Gokhan Turer, and David Talby.
Proceedings of The Seventh Workshop on Social Media Mining for Health Applications, Workshop & Shared Task, pages 44–47, Gyeongju, Republic of Korea.
Social media has become a major source of information for healthcare professionals but due to the growing volume of data in unstructured format, analyzing these resources accurately has become a challenge. In this study, we trained health related NER and text classification models on different datasets published within the Social Media Mining for Health Applications (#SMM4H 2022) workshop1.
We utilized transformer based Bert for Token Classification and Bert for Sequence Classification algorithms as well as vanilla NER and text classification algorithms from Spark NLP library during this study without changing the underlying DL architecture. The trained models are available within a production-grade code base as part of the Spark NLP library; can scale up for training and inference in any Spark cluster; has GPU support and libraries for popular programming
languages such as Python, R, Scala and Java.