Healthcare providers and well-established players in the healthcare space possess vast amounts of unstructured patient-level data. This data has tremendous value, yet it stays primarily untapped due to the sensitivity and legal and compliance requirements for working with them. However, if the data were de-identified, it would allow the building of a whole new sector of the economy filled with innovative solutions benefitting all.
The advent of new solution builders using unstructured, patient-level data
With the increasing power of machine learning, the rise of artificial intelligence, and big data, there is ever-increasing potential to build innovative solutions in healthcare and pharma. Many players in the field are eager to take on the challenge – including healthcare and pharma startups, new business lines of established companies, and even organically growing transformational pharma and healthcare corporations.
Many of those solutions use machine learning models or require a database of patient-level data to build and run the system. The solution builders’ business models allow for profit-sharing ultimately arising from the operation of the solution back to the providers of the unstructured patient-level data, i.e., they are ready to pay for the data.
One solution does not fit all. De-Id is per project challenge
The data needs to be de-identified first before further processing. The need come from the sensitivity of the data. It is regulated in every country, usually with strict requirements – e.g., HIPAA de-identification Privacy Rule in the US and GDPR across the EU.
Each de-identification project requires a proper strategy, as the de-identification method needs to be aligned with the intended data use. The stricter deidentification rules lower the risk of re-identification. However, it also decreased the usability and value of the data. E.g., if the relative time between events needs to be preserved, the date in the year cannot be simply removed. If a subsequent analysis is related to infants, the age in days may be required. However, keeping this information in the data set increases the risk of re-identification. On the other hand, even the strictest de-identification rules do not entirely remove re-identification risk. There is always some residual risk present.
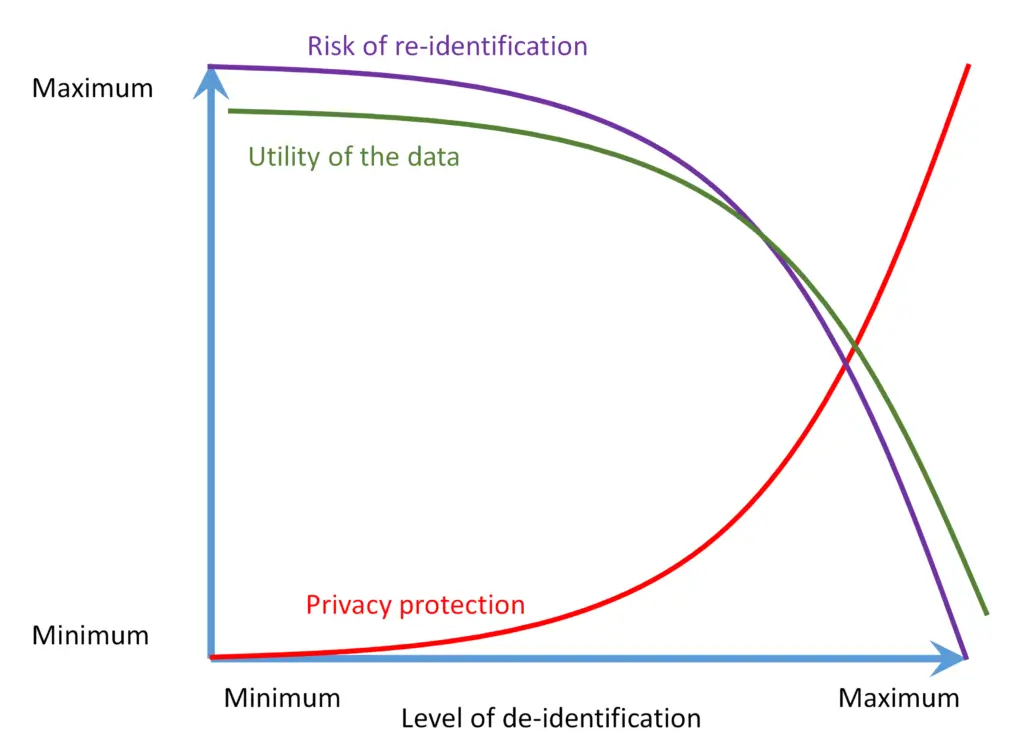
Figure 1: Privacy / utility trade-off
Less accurate than you expect
Manual removal of protected health information (PHI) from the data set is never flawless. Achieving high accuracy requires experience and good organization of the annotation team:
- Well trained annotators
- High-quality annotation guidelines, with proper specification of de-identification rules
- Well-controlled process for consistency across annotators and documents
- Four eyes principle – one team of annotators working, while the second team approves their work
- Process for sharing and feedback on annotation differences
The recall figures in the literature are lower than one may think. See, e.g., [Douglas at al 2004]:
| Process | Min | Max | Mean |
| 1 reviewer | 63% | 94% | 81% |
| 2 reviewers | 89% | 98% | 94% |
Other figures published in the literature, e.g. [Dorr at al 2006], report 96% recall for single review performance. However, ground truth there is defined as two reviewers’ performance, which, as noted above, may be significantly different from the real ground truth.
Manual deidentification is prohibitively expensive for large data sets. The de-identification is traditionally limited to small or mid-size data samples. [Douglass et al 2004] reported the team with an average cost of $83 per hour total compensation, including bonus, processing 135 notes per hour of an average length of 130 words. It costs $0.61 per note (or $2.34 per 500 words page).
Thus, large data sets consisting of millions of records cannot be manually de-identified due to considerable labor required at prohibitive cost.
Power of automatic de-identification of unstructured patient-level data and John Snow Labs automatic de-identification services
An increase in natural language processing (NLP) in healthcare allows automatically getting de-identified health information from unstructured patient-level data in many languages. The automated system can perform on par with humans if well-tuned for the case in consideration.
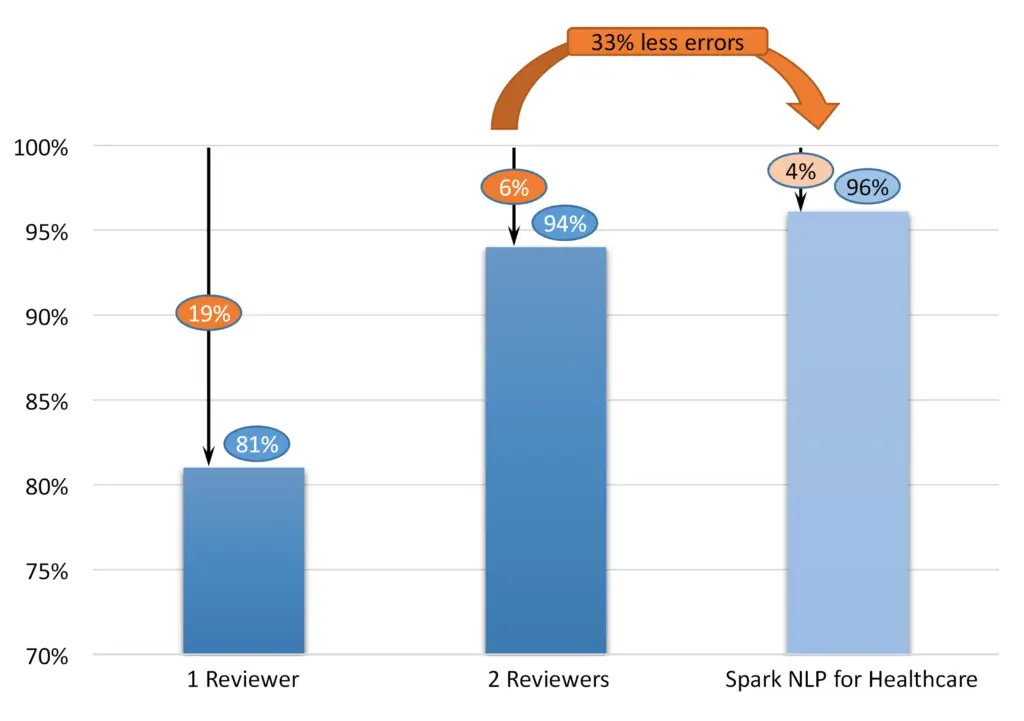
Figure 2: Performance of human reviewers vs. NLP
John Snow Labs’ automatic data de-identification software provides custom de-identification required for the monetization of the data. The service is based on Spark NLP for the Healthcare library [John Snow Labs]. The library is built on top of the Spark big data framework, allowing the process of millions of records on large Spark or Databricks clusters. The de-identification solution can be delivered as an end-to-end system or an IT library with optional professional services. In 2022, John Snow Labs established a new state-of-the-art record on the n2b2 standard de-identification benchmark [Kocaman and Talby 2022], achieving an F1 score of 96.1%. John Snow Labs keep improving the solution regularly, publishing the release every few weeks. Please, note the critical difference between 96% and 94% – means decreasing the error rate from 6% to 4%, i.e., a decrease by 33%. Healthcare Natural Language Processing by John Snow Labs powers many industrial solutions. See, e.g., [Piotrowski, Dobes 2022], where IQVIA reported an F1 score of 95% for the German language.
Once the data are de-identified, the owners can share the dataset with the innovative solution builders. Without large-scale automatic data deidentification, the solution cannot be built. Thus automatic de-identification technology is an enabler of innovative AI and NLP solutions in healthcare.
References:
Neamatullah et al 2008, Ishna Neamatullah, Margaret M Douglass, Li-wei H Lehman, Andrew Reisner, Mauricio Villarroel, William J Long, Peter Szolovits, George B Moody, Roger G Mark & Gari D Clifford: Automated de-identification of free-text medical records. BMC Medical Informatics and Decision Making volume 8, Article number: 32 (2008)
Douglas et al 2005, Douglass, M. J., Cliffford, G. D., Reisner, A. et al. De-identification algorithm for free-text nursing notes. In CinC (2005)
Douglass at al 2004, M. Douglass, G. D. Clifford, A. Reisner, G. B. Moody and Mark RG, “Computer-assisted de-identification of free text in the MIMIC II database,” Computers in Cardiology, 2004, 2004, pp. 341-344, doi: 10.1109/CIC.2004.1442942.
Dorr at al, 2006, Dorr, D & Phillips, W & Phansalkar, Shobha & Sims, S & Hurdle, J. (2006). Assessing the Difficulty and Time Cost of De-identification in Clinical Narratives. Methods of information in medicine. 45. 246-52. 10.1055/s-0038-1634080.
John Snow Labs, Spark NLP for Healthcare
Kocaman and Talby 2022, Kocaman, V., and Talby, D. “Accurate clinical and biomedical Named entity recognition at scale.” Software Impacts (2022): 100373. 10.1016/j.simpa.2022.100373
Piotrowski, Dobes 2022, Using Spark NLP to De-Identify Doctor Notes in the German Language. Healthcare NLP Summit 2022. https://www.nlpsummit.org/using-spark-nlp-to-de-identify-doctor-notes-in-the-german-language/