Entity Resolution is the process of predicting UMLS codes for medical concepts. While processing medical text, this process relies heavily on the concepts identified by NER models. NER models at first identify medical models, which are then fed to the Entity Resolver models to get UMLS codes. In most cases, these concepts/chunks are very concise.
For example, in the example below, the word “fracture” is identified as a problem, while the location of the problem is “leg” with further segregation of laterality.

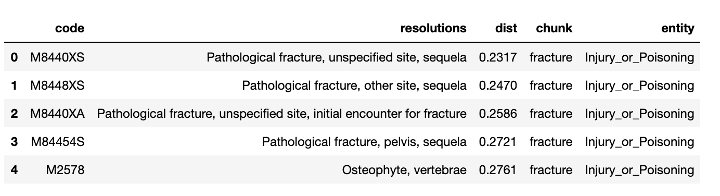
If we just use the chunk “fracture” to predict code for the problem (the previous approach is used), we’d get something like this:
 Top 5 predicted codes for the given chunk – These are the results of the previous approach used so far.
Top 5 predicted codes for the given chunk – These are the results of the previous approach used so far.
In theory, the resolver model is doing its job just fine, as we are asking it to predict code for just “fracture”, but more precise results can be achieved if the enrich it with the body part.
SentenceChunkEmbeddings Annotator
This problem can be tackled with the new annotator called “SentenceChunkEmbeddings”. This annotator would take weighted average of the embedding vector of each chunk and the entire sentence, producing embeddings that is enriched with contextual information, which can directly be used in the Entity Resolver model. The following code explains how to use it inside a pipeline:
Code: python
documenter = DocumentAssembler()
.setInputCol("text")
.setOutputCol("documents")
sentence_detector = SentenceDetector()
.setInputCols("documents")
.setOutputCol("sentences")
tokenizer = Tokenizer()
.setInputCols("sentences")
.setOutputCol("token")
embeddings = WordEmbeddingsModel()
.pretrained("embeddings_clinical", 'en', 'clinical/models')
.setInputCols(["sentences","token"])
.setOutputCol("embeddings")
ner = MedicalNerModel()
.pretrained("ner_jsl_greedy", "en", "clinical/models")
.setInputCols("sentences", "token","embeddings")
.setOutputCol("ner")
ner_converter = NerConverter()
.setInputCols("sentences", "token", "ner")
.setOutputCol("ner_chunk")
sentence_chunk_embeddings = BertSentenceChunkEmbeddings
.pretrained("sbiobert_base_cased_mli", "en", "clinical/models")
.setInputCols(["sentences", "ner_chunk"])
.setOutputCol("sentence_chunk_embeddings").setChunkWeight(0.5) #default : 0.5
resolver = SentenceEntityResolverModel.pretrained('sbiobertresolve_icd10cm', 'en', 'clinical/models')
.setInputCols(["ner_chunk", "sentence_chunk_embeddings"])
.setOutputCol("resolution")
pipeline = Pipeline().setStages(
[documenter,
sentence_detector,
tokenizer,
embeddings,
ner,
ner_converter,
sentence_chunk_embeddings,
resolver
])
Pipeline_model = pipeline.fit(spark.createDataFrame([['']]).toDF("text"))
````
Running the new pipeline on the same piece of text generates many accurate results as shown below:
 Top 5 codes – These results are achieved with a default chunk weight of 0.5
Top 5 codes – These results are achieved with a default chunk weight of 0.5
As we can see the resultant codes are now much more accurate. Although the chunk is still the same, since the embeddings are not enriched with more useful context, the model was able to predict codes for fracture of left fibula, which is a lot more precise and useful than simple fracture.
Annotator Parameters
The parameter “setChunkWeight” can be used to control the contribution of each embedding. A value of 1 will give full weightage to the chunk – this can be used when the chunk is enriched with the required information already, or if the sentence is too long and complex with multiple entities. While a value of 0 would give full weightage to the overall sentence – this is helpful when the sentences are small and less complex. By default, the value is set at 0.5 to make an equal representation of the sentence and the chunk.
Other uses
The introduction of this annotator also eliminates the need of following the old process of converting NER chunks to documents, and generating embeddings manually before feeding them to the resolver. Now users can just use this single annotator to achieve the same functionality by setting the chunk weight value to 1. The modified pipeline is explained in the diagram below:
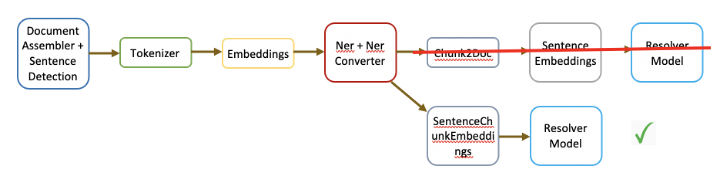
Further Resources
Google Colab notebook for this post is available here.
Check out all existing functionalities are available in Spark NLP for Healthcare here.

























 Top 5 predicted codes for the given chunk – These are the results of the previous approach used so far.
Top 5 predicted codes for the given chunk – These are the results of the previous approach used so far. Top 5 codes – These results are achieved with a default chunk weight of 0.5
Top 5 codes – These results are achieved with a default chunk weight of 0.5




