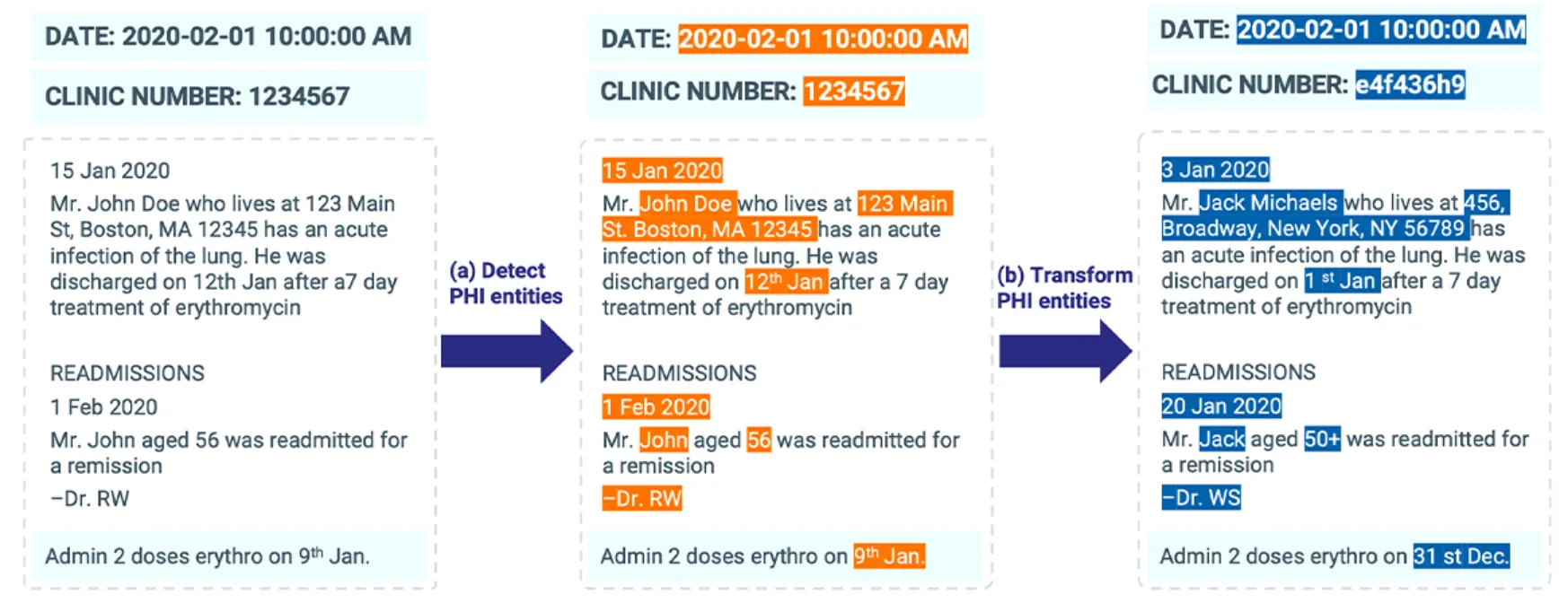
GLiNER and OpenPipe Shine on General Texts but Miss Over 50% of Clinical PHI — Compared to Less Than 5% Misses by Solutions Like John Snow Labs
It’s often assumed that all PII and PHI entities are the same, meaning de-identification tools should work equally well across all unstructured text. While this seems logical in theory, each domain-specific dataset has distinct characteristics, requiring customized approaches for effective PII detection. In this blog post, we highlight how LLM-based de-identification models, such as GLiNER PII and OpenPipe’s PII-Redact, excel in general text — achieving macro-average F1 scores of 0.62 for GLiNER and 0.98 for OpenPipe — but face significant performance drops on clinical datasets, where their scores fall to 0.41 and 0.42 respectively.
What is PII/PHI and Why is Deidentification Important?
Personally Identifiable Information (PII) and Protected Health Information (PHI) refer to any data that can be used to identify an individual, such as names, addresses, phone numbers, social security numbers, or medical records. In sectors like healthcare, finance, and law, safeguarding this sensitive information is not just a best practice — it’s a legal requirement under regulations like HIPAA (in the U.S.) and GDPR (in the EU).
Deidentification is the process of removing or masking PII/PHI from datasets to protect individuals’ privacy. It is crucial when sharing data for research, analysis, or machine learning model development. Effective deidentification ensures compliance with privacy laws while enabling valuable insights to be drawn from real-world data — especially in healthcare, where access to clinical text can drive innovation in patient care and medical research.
In this blog post, we compare the PII/PHI entity extraction performance of two open-source tools used for PII detection: OpenPipe’s PII-Redact and GLiNER PII model. The evaluation is conducted on two different datasets: the publicly available AI4Privacy dataset, which contains samples from a variety of domains, and a real-world clinical dataset, labeled by domain experts specifically for healthcare-related PHI. Our goal is to assess how effectively these tools identify PII/PHI in both general unstructured text and domain-specific clinical narratives.
Tools Compared
What is PII-Redact?
PII-Redact is an open-source Python library designed for detecting and redacting Personally Identifiable Information (PII) in text. It efficiently extracts sensitive entities such as age, date, email address, personal ID, and person name, among others. You can see all the list of entities from the github.
The library utilizes Llama 3.2 1B LLMs to identify PII in unstructured text. Unlike traditional rule-based or regex-based solutions, PII-Redact benefits from large language models (LLMs), enabling it to adapt to various data formats and contexts. Users can customize its behavior with different levels of strictness, choosing whether to replace, redact, or mask detected PII elements.
After installing the PII-Redact library, you can easily obtain predictions by running the following command:
from pii_redaction import tag_pii_in_documents, clean_dataset, PIIHandlingMode # Process text documents documents = ["My name is John Doe and my email is john.doe@example.com"] # Tag PII (default mode) tagged_documents = tag_pii_in_documents(documents, mode=PIIHandlingMode.TAG)
The output appears as follows:
'My name is John Doe and my email is john.doe@example.com'
PII-Redact’s tag_pii_in_documents method marks PHI entities using HTML tags, embedding labels within them. To compare these results with the ground truth, we needed to process the output by removing HTML tags and extracting the relevant start and end indices, entity text (chunk), and corresponding label. This required additional post-processing to align the detected entities with the labeled datasets.
What is GLiNER PII Model?
GLiNER PII is a Named Entity Recognition (NER) model designed to identify a wide range of personally identifiable information (PII) within text. Utilizing a bidirectional transformer encoder similar to BERT, GLiNER offers a practical alternative to traditional NER models limited to predefined entities and to large language models (LLMs) that may be resource-intensive. This model is capable of recognizing various PII types, including but not limited to names, addresses, phone numbers, email addresses, and social security numbers.
After installing the GLiNER, you can utilize it by selecting the model as follows:
from gliner import GLiNER
model = GLiNER.from_pretrained("urchade/gliner_multi_pii-v1")
labels = ['IDNUM', 'LOCATION', 'DATE', 'AGE', 'NAME', 'CONTACT']
entities = model.predict_entities(text, labels)
Evaluation Criteria
In this benchmark study, we adopted two different approaches to assess accuracy across both general-domain unstructured text and clinical datasets:
A. Entity-Level Evaluation: Since de-identifying PII/PHI data is a critical task, we evaluated how well entity extraction performed in the datasets, regardless of their specific labels in the ground truth. The detection outcomes were categorized as:
- full_match: The entire entity was correctly detected.
- partial_match: Only a portion of the entity was detected.
- not_matched: The entity was not detected at all.
For example:
Text: “Patient John Doe was admitted to Boston General Hospital on 01/12/2023.”
Ground Truth Entity: John Doe (NAME)
Predicted Entity: John Doe (NAME) ==> full_match
Predicted Entity: John==> partial_match
If predictions don’t have any match with “John Doe”, the result is “not_matched”.
B. Token-Level Accuracy: The datasets were tokenized, and the ground truth labels for each token were compared against the predictions. To evaluate performance, classification reports were generated, analyzing precision, recall, and F1 scores.
This dual approach comprehensively evaluated the performance of the tools in de-identifying PHI data.
Benchmark #1: AI4Privacy PII-Masking-300K Dataset
Dataset
The PII-Masking-300K dataset, created by AI4Privacy, is a large-scale dataset designed for training and evaluating Personally Identifiable Information (PII) masking models. It contains ~220.000+ text samples with PII annotations and corresponding masked versions, making it a valuable resource for privacy-preserving NLP applications.
Key Features:
- Size: ~220.000+ text samples
- Supported Languages: 6 languages (English (UK, USA), French (France, Switzerland), German (Germany, Switzerland), Italian (Italy, Switzerland) Dutch (Netherlands), Spanish (Spain) in total with strong localisation in 8 jurisdictions.
- Annotations: Includes PII-labeled data with entities such as names, addresses, emails, phone numbers, and other sensitive identifiers
- Masked Versions: Each sample has a corresponding deidentified version, where PII has been replaced with placeholders or redacted
- Diverse Sources: Covers a wide range of text formats, ensuring robustness for real-world PII detection and masking tasks
- Use Cases: Ideal for training, benchmarking, and evaluating PII detection models in healthcare, finance, and legal sectors
This dataset serves as a benchmarking resource for developing privacy-focused AI models, ensuring compliance with data protection regulations like GDPR and HIPAA.
In this study, we selected 100 English samples from the dataset and conducted benchmarks on them.
Results
Entity-Level Evaluation:
The results from aligning the predictions of OpenPipe’s PII-Redact and the GLiNER PII model with the ground truth entities are presented below:
Based on these results, the matching percentages representing the match rates for the de-identification tools are plotted as shown below:
Token-Level Evaluation:
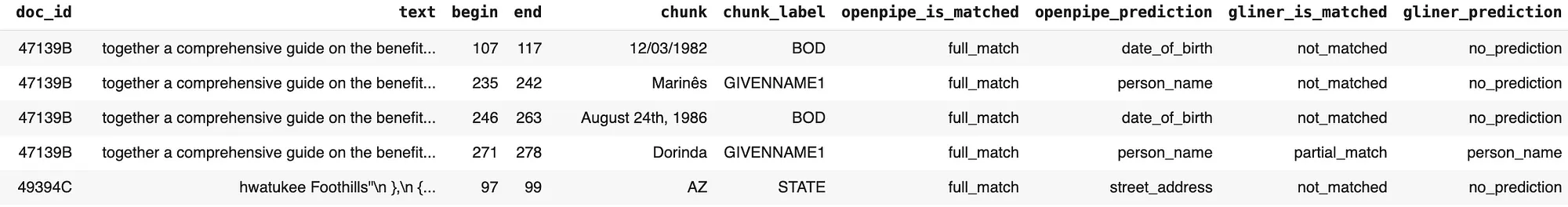
The resulting data frame, created by tokenizing the ground truth–annotated text and attaching the corresponding ground truth labels alongside the predictions from OpenPipe’s PII-Redact and the GLiNER PII model for each token, is shown below:
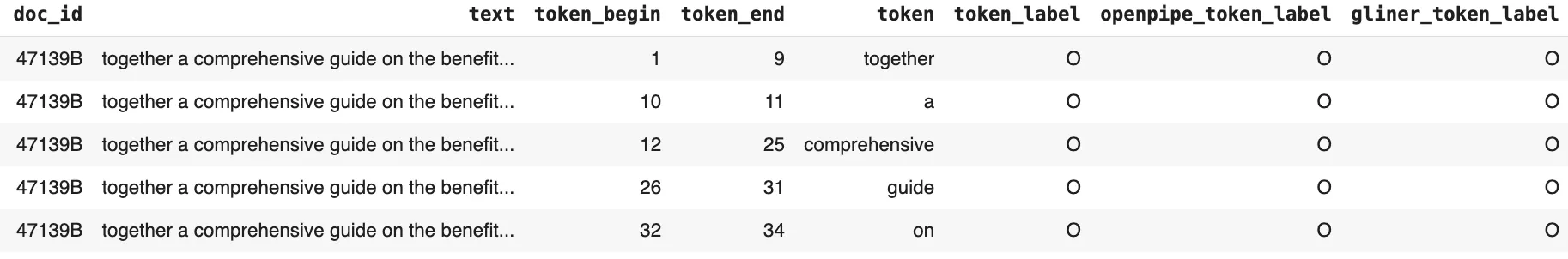
Token Level Results Dataframe
Since the prediction labels and ground truth labels differ, we mapped the ground truth labels to the more generic prediction labels to ensure a fair comparison. Additionally, some prediction labels — such as organization_name—were not present in the ground truth dataset, so we excluded them from the token-level evaluation.
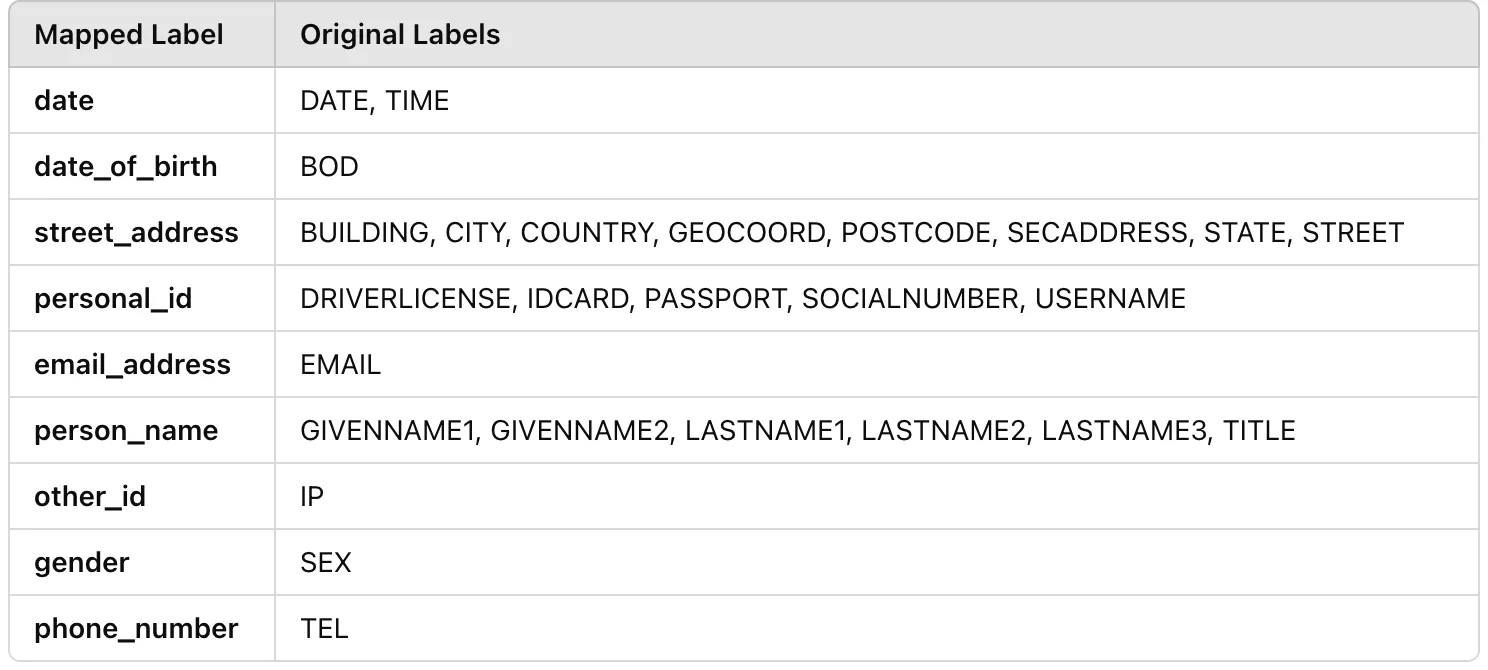
Label Mapping Table
Based on the token-level results, the classification report showing the comparison between the OpenPipe’s PII-Redact predictions and the AI4Privacy sample dataset is as follows:
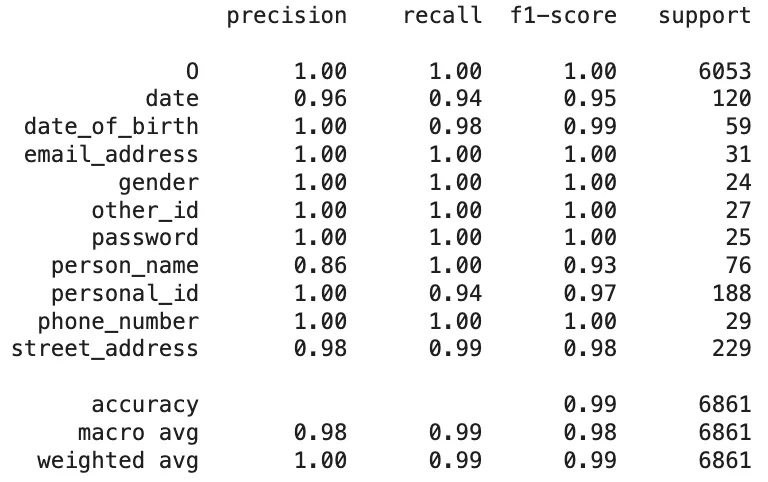
Classification Report of The PII-Redact’s Predictions On AI4Privacy Dataset
For a fair comparison, we aligned the labels returned by PII-Redact with those used in the GLiNER PII Model. Based on the token-level evaluation, the following classification report presents a comparison between the GLiNER PII predictions and the AI4Privacy sample dataset:
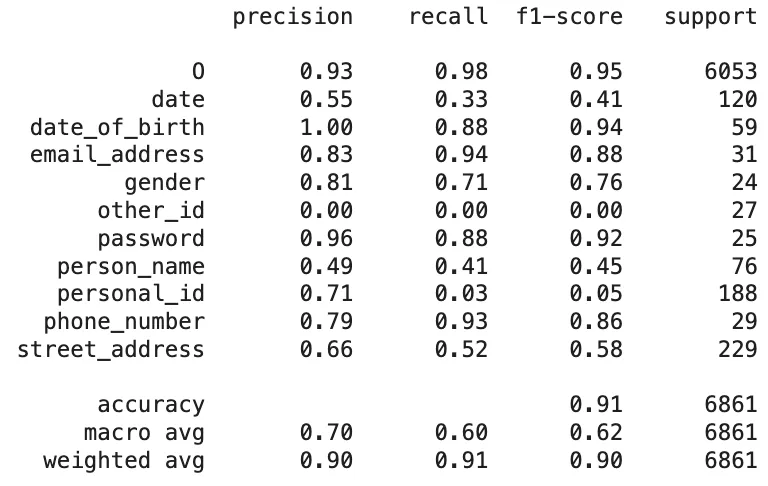
Classification Report of The GLiNER PII Predictions On AI4Privacy Dataset
PHI Entity Prediction Comparison:
The de-identification task is crucial in ensuring the privacy of sensitive data. In this context, the primary focus is not on the specific labels of PHI entities but on whether they are successfully detected. When evaluating the results based on the classification of entities as PHI or non-PHI, the outcomes for PHI entity detection are as follows:
PII-Redact’s PHI Entity Detection Scores On AI4Privacy Dataset
GLiNER PII’s PHI Entity Detection Scores On AI4Privacy Dataset
Benchmark #2: Real-World Clinical Dataset
Dataset
For this benchmark, we used 48 open-source documents annotated by domain experts from John Snow Labs. The annotations focused on extracting IDNUM, LOCATION, DATE, AGE, NAME, and CONTACT entities, as these represent key personal information frequently targeted for deidentification in healthcare settings. These entities are closely tied to an individual’s identity and often require removal or anonymization to comply with regulations like HIPAA and GDPR. By centering the benchmark on these categories, we ensure that the evaluation directly addresses real-world deidentification challenges, where accurately detecting and obscuring such sensitive data is critical.
The PHI entity extraction results of major cloud vendors and John Snow Labs on this dataset are detailed in the paper “Can Zero-Shot Commercial APIs Deliver Regulatory-Grade Clinical Text DeIdentification?”. For a deeper dive into the study and its findings, you can also refer to our related blog post, Comparing Medical Text De-Identification Performance.
Results
We found that both tools performed poorly on the clinical dataset, and the GLiNER model has a max_len restriction. Given that the clinical texts in the dataset vary in length, with some being quite lengthy, we used John Snow Labs’ DocumentSplitter to divide the documents into smaller splits. Predictions were then obtained from both tools based on these splits to ensure a fair comparison.
Entity-Level Evaluation:
The results from comparing the predictions made by OpenPipe’s PII-Redact and GLiNER PII models with the clinical ground truth entities are as follows:
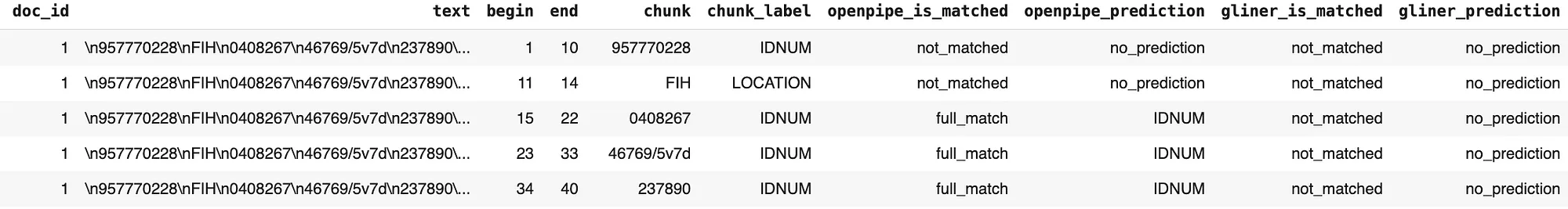
Matching Status On Clinical Dataset
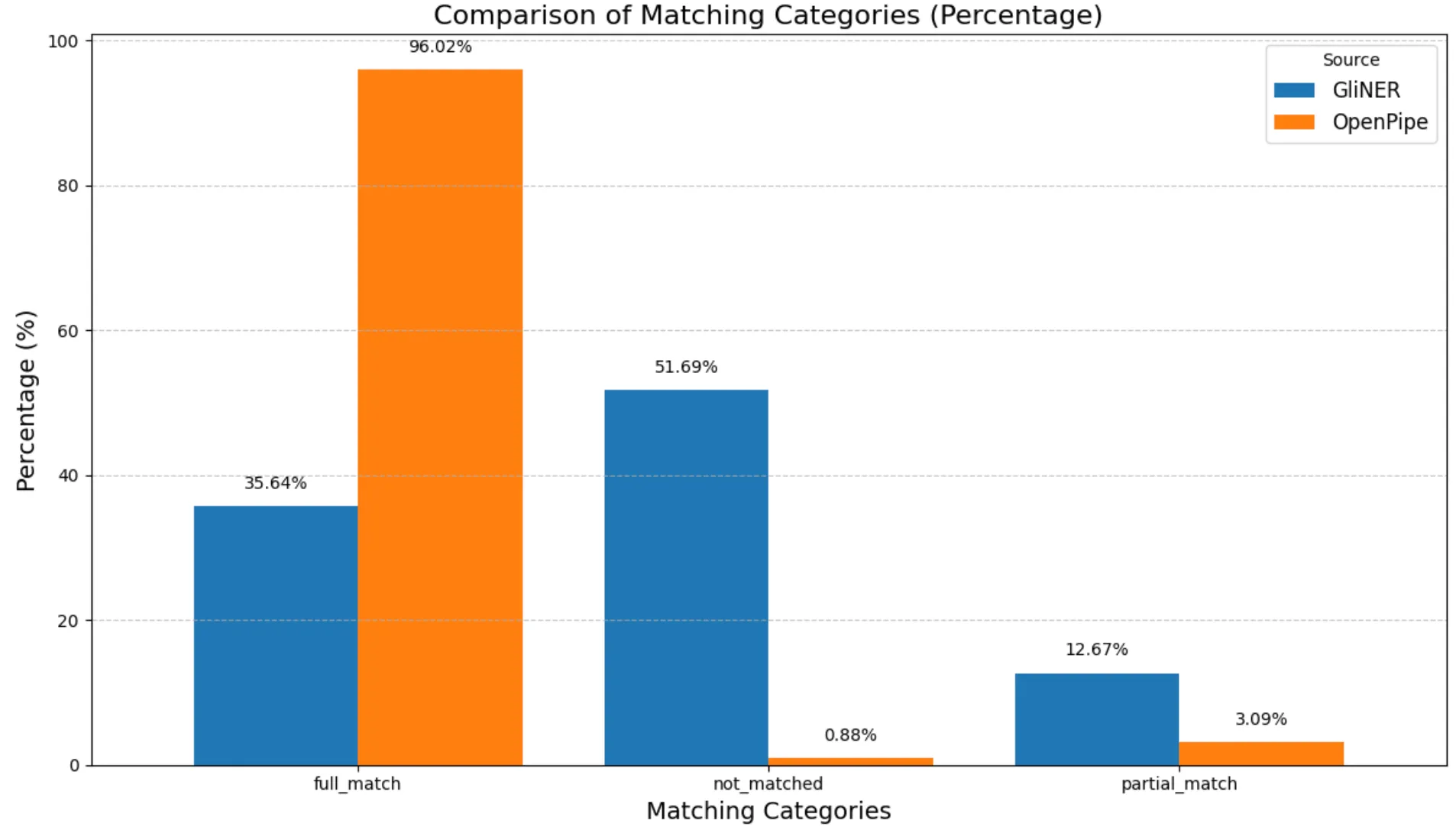
Based on these results, the match percentages for the de-identification tool on the clinical dataset, representing the rates of prediction accuracy, are plotted below:
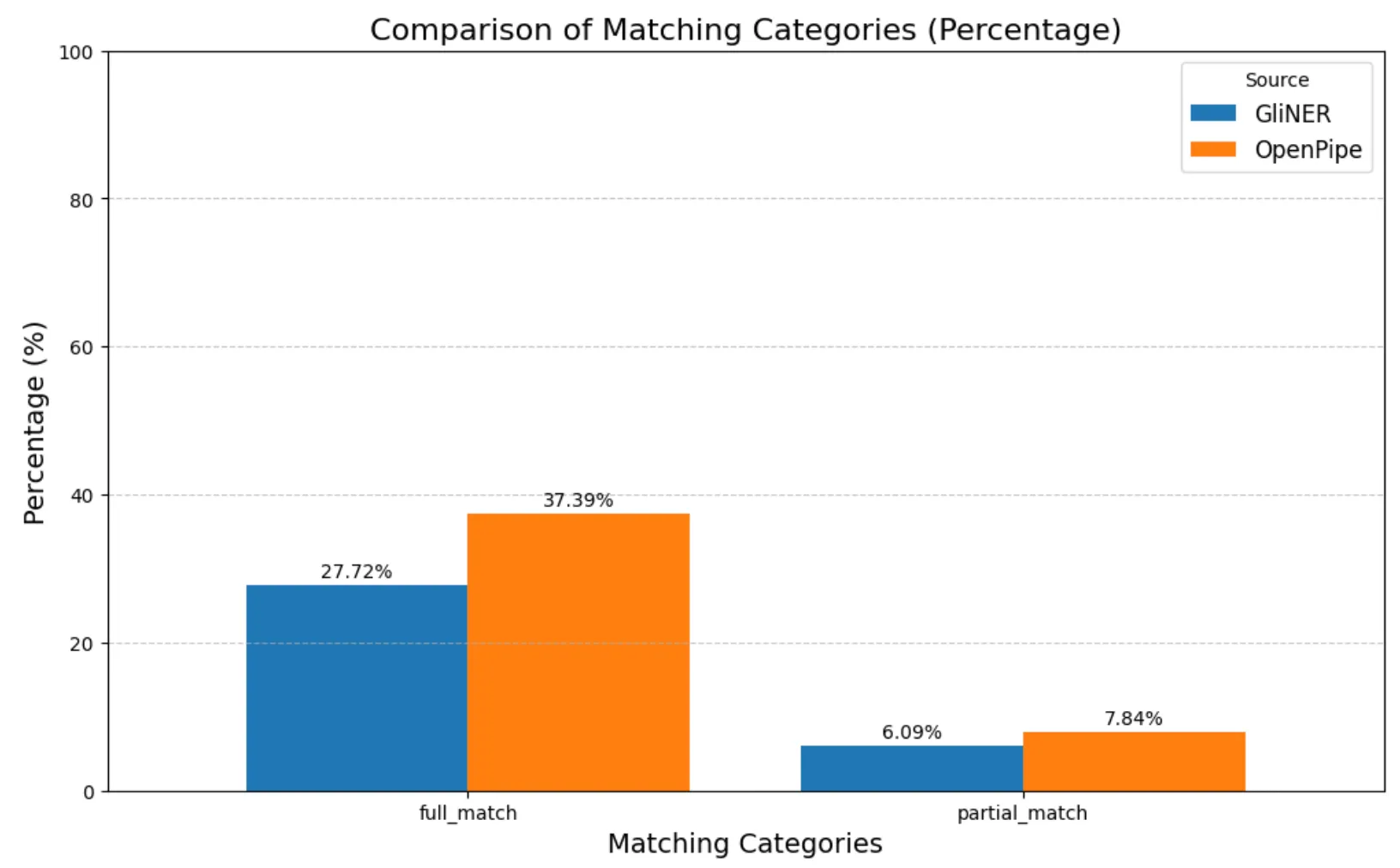
Entity Match Results In Percentage On Clinical Dataset
Token-Level Evaluation:
The resulting data frame, created by tokenizing the ground truth text and appending the corresponding ground truth labels, along with the prediction labels from OpenPipe PII-Redact and the GLiNER PII model for each token, is as follows:
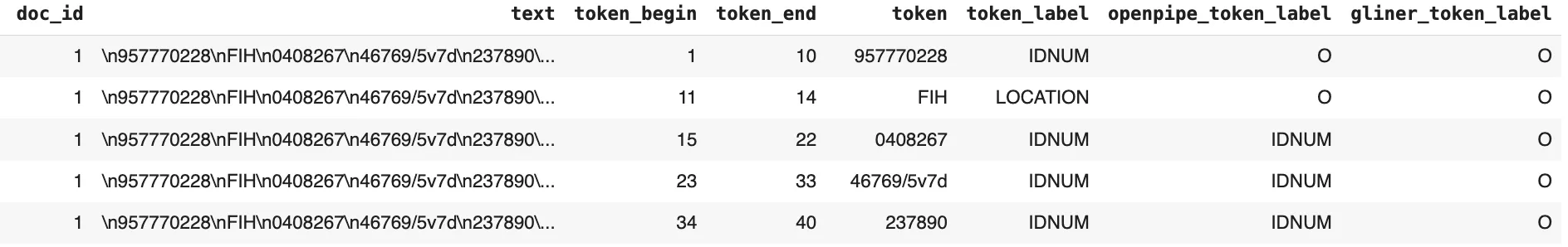
Token Level Results Dataframe
Following the same approach as with the AI4Privacy dataset, we mapped the PII-Redact prediction labels to the more generic corresponding ground truth labels (IDNUM, LOCATION, DATE, AGE, NAME, CONTACT) as shown below. Entities that could not be matched to any ground truth labels (e.g., organization_name, gender) were excluded from the predictions prior to evaluation. For the GLiNER model, we had already assigned these ground truth labels to the predictions.
Note: The predictions did not include any other labels, including AGE.

PII-Redact Label Mapping Table
Based on the token-level results, the classification report showing the comparison between the OpenPipe’s PII-Redact predictions and the clinical ground truths are as follows:
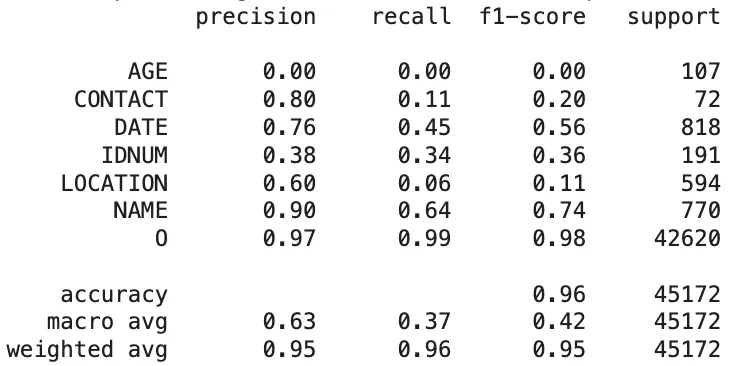
Classification Report of The PII-Redact’s Predictions On Clinical Dataset
Here are the GLiNER PII model token level evaluation results:
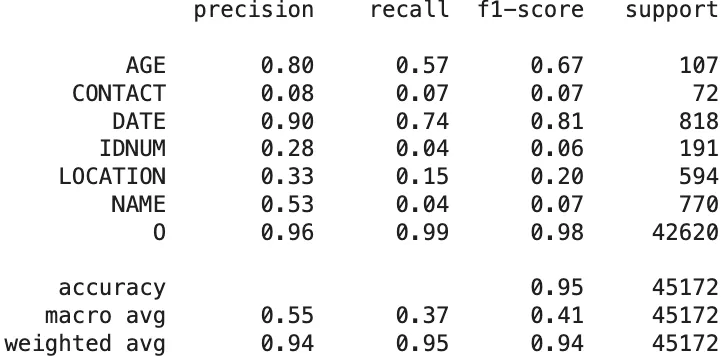
Classification Report of The GLiNER PII Model Predictions On Clinical Dataset
PHI Entity Prediction Comparison:
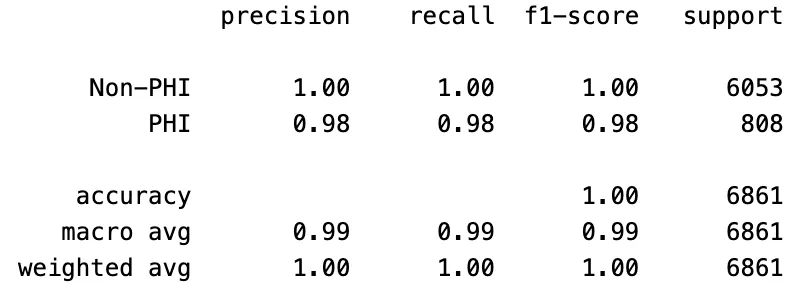
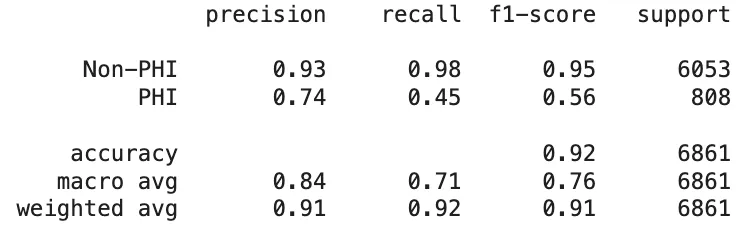
As mentioned earlier, the de-identification task plays a vital role in safeguarding the privacy of sensitive information. The key focus here is not necessarily on the specific labels of PHI entities, but rather on whether these entities are correctly detected. When evaluating the results based on the classification of entities as PHI or non-PHI, the outcomes for PHI entity detection are as follows:
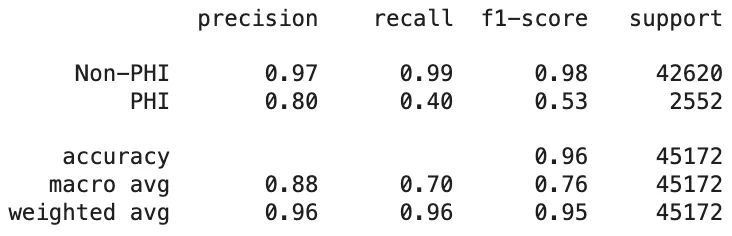
PII-Redact’s PHI Entity Detection Scores On Clinical Dataset
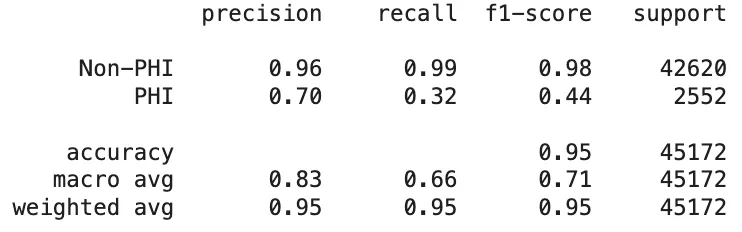
PII-Redact’s PHI Entity Detection Scores On Clinical Dataset
Performance of Zero-Shot Commercial APIs on Clinical Datasets
De-identifying Protected Health Information (PHI) in clinical datasets is crucial for safeguarding patient privacy and ensuring compliance with regulations such as HIPAA and GDPR. While tools like OpenPipe’s PII-Redact and the GLiNER PII model have shown limited effectiveness in this area, a recent study evaluated the performance of several zero-shot commercial APIs — including John Snow Labs’ Healthcare NLP, OpenAI’s GPT-4o, Anthropic’s Claude 3.7 Sonnet, Azure Health Data Services, and Amazon Comprehend Medical — on the same clinical dataset.
The findings revealed that John Snow Labs outperformed the other tools in accurately detecting PHI entities within clinical texts. This tool demonstrated superior precision and recall rates, highlighting its effectiveness in handling sensitive medical data. In contrast, the other APIs exhibited varying degrees of performance, with some showing limitations in identifying certain PHI categories.
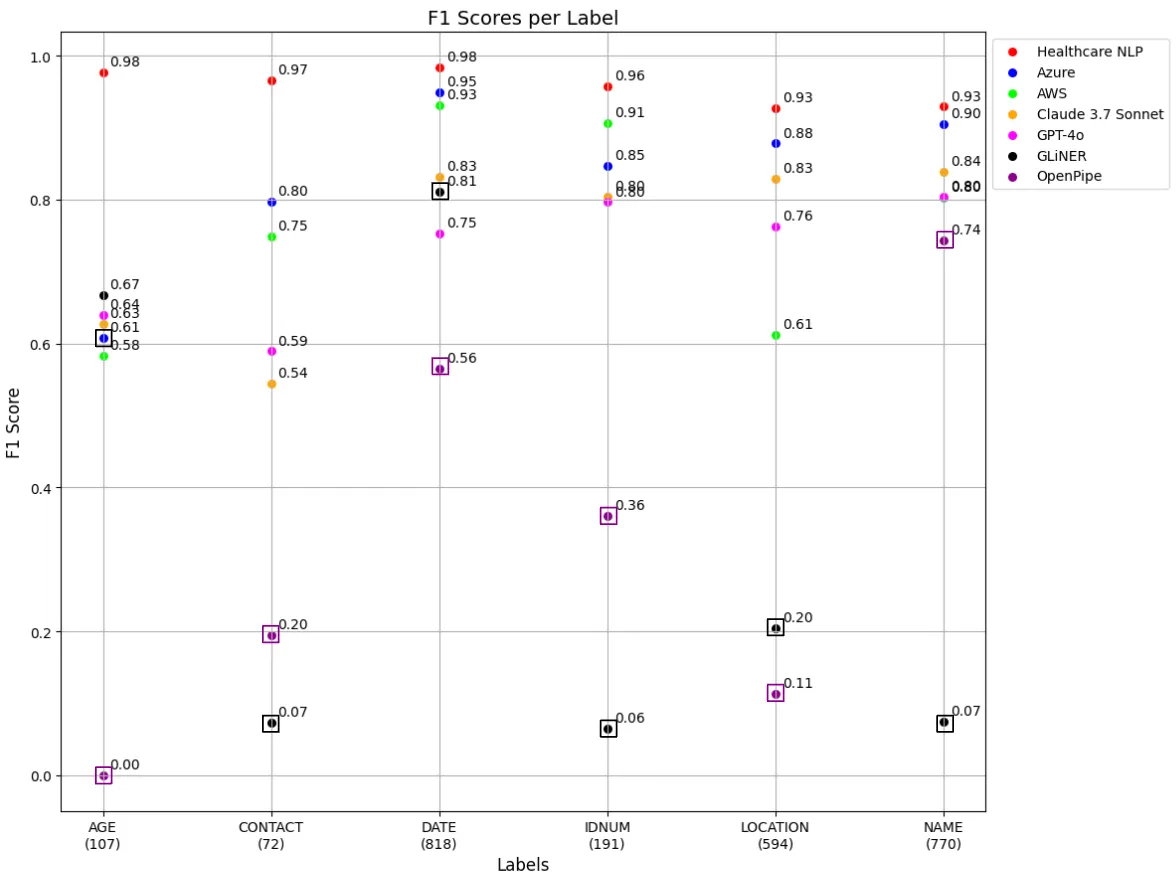
F1 Score Table For Each Label
A key differentiator for John Snow Labs is its adaptability. Unlike the other API-based, black-box cloud solutions, John Snow Labs offers a highly customizable de-identification pipeline that can be tailored to specific requirements. Additionally, it can be deployed locally, providing organizations with greater control over data processing without the need for an internet connection.
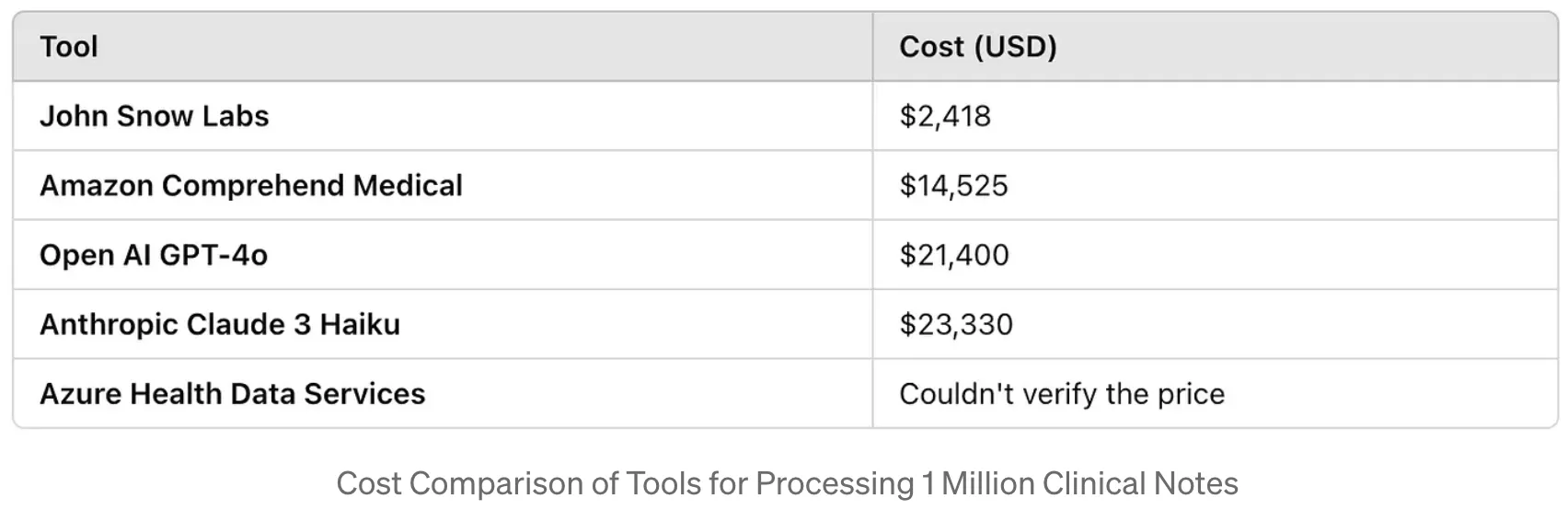
Cost analysis further underscores John Snow Labs’ advantages. When processing large volumes of clinical notes, it proves more cost-effective than other API-based services, which often incur higher expenses due to per-request pricing models. This affordability, combined with its high performance and customization capabilities, positions John Snow Labs’ Healthcare NLP as a leading solution for de-identifying PHI in clinical datasets.
Findings and Discussion
In this study, we benchmarked OpenPipe’s PII-Redact and the GLiNER PII model on two distinct datasets: a general-purpose dataset (AI4Privacy) and a healthcare-specific clinical dataset. The results highlight how these tools perform across different domains, revealing both their strengths and limitations.
Performance on AI4Privacy Dataset
Entity-Level Evaluation:
- OpenPipe PII-Redact demonstrated exceptional accuracy, with 96.02% full match rate, and only 0.88% of entities not matched, reflecting its robustness in identifying and labeling entities precisely. Partial matches were minimal at 3.09%.
- GLiNER PII showed significantly lower performance, with only 35.64% full match rate, and a high 51.69% not matched rate, indicating difficulties in fully capturing the correct entity boundaries. The partial match rate was 12.67%, suggesting moderate ability to recognize some PII components but often incompletely.
Token-Level Evaluation:
- OpenPipe achieved an overall accuracy of 99%, with a macro-average F1-score of 0.98 and near-perfect scores for most entity types. Key entity types such as
email_address,gender,password,phone_number, andstreet_addressall had F1-scores of 0.98 to 1.00. Only slight dips were observed inperson_name(F1-score: 0.93) andpersonal_id(0.97), but even these remained strong. - GLiNER, while still functional, lagged behind significantly. Its overall accuracy was 91%, with a macro-average F1-score of just 0.62. Several categories like
personal_id(F1-score: 0.05),other_id(0.00), anddate(0.41) were particularly problematic. Althoughemail_address(0.88) andphone_number(0.86) scored reasonably well, the general inconsistency across entities limited its reliability for precise de-identification.
Performance on Clinical Dataset
Entity-Level Evaluation:
- OpenPipe performed moderately better with a full match rate of 37.39%, compared to GLiNER’s 27.72%, indicating a higher ability to correctly extract full PHI entities.
- Not matched rates remained high for both tools, reflecting the overall challenge of PHI recognition in clinical text: 54.77% for OpenPipe and 66.19% for GLiNER.
- Partial matches were relatively low in both models, with 7.84% for OpenPipe and 6.09% for GLiNER, indicating limited cases where the tools captured entity fragments but missed the full span.
Token-Level Evaluation:
- OpenPipe’s PII-Redact demonstrated a good performance on structured entities such as
NAME(F1-score: 0.74) andDATE(0.56), indicating a solid ability to detect more standardized PHI types. However, its effectiveness dropped significantly for other critical categories, includingAGE(0.00) (no AGE prediction observed),CONTACT(0.20),IDNUM(0.36), andLOCATION(0.11), highlighting difficulties in handling diverse and context-sensitive identifiers. The macro-average F1-score was 0.42, pointing to inconsistent performance across entity types despite some high scores. - GLiNER outperformed OpenPipe in specific categories such as AGE (F1-score: 0.67) and
DATE(0.81), but underperformed on key fields likeNAME(0.07),CONTACT(0.07), andIDNUM(0.06), where the detection rates were particularly low. Performance onLOCATIONwas also limited, with an F1-score of 0.20, indicating continued difficulty with geographical identifiers. The macro-average F1-score for GLiNER stood at 0.41, closely mirroring OpenPipe, but with sharp variations between categories that reflect an overall lack of consistency across PHI types.
Comparative Analysis with Commercial APIs:
A comparative analysis involving zero-shot commercial APIs — such as John Snow Labs, OpenAI, Anthropic Claude, Azure Health Data Services, and Amazon Comprehend Medical — was performed using the same clinical dataset. Among these, John Snow Labs consistently delivered the best performance in identifying PHI entities, achieving the highest scores in both entity-level and token-level evaluations. Its results surpassed not only those of the open-source tools but also other commercial solutions in the task of clinical text de-identification.
Conclusion
In conclusion, OpenPipe’s PII-Redact demonstrated strong performance on unstructured datasets spanning various domains, achieving high accuracy in both entity- and token-level evaluations. Its ability to identify a wide range of PII entities with near-perfect precision and recall makes it a solid choice for general-purpose de-identification tasks. On the other hand, the GLiNER PII model fell short in delivering reliable results for PII extraction. Despite being lightweight and easy to use, GLiNER struggled to maintain consistent accuracy across different entity types and failed to meet the performance expectations for a task as critical as PII detection.
That said, while LLM-based open-source tools like GLiNER and OpenPipe show promise in handling certain types of PII entity detection across multiple domains, they fall short when applied to domain-specific datasets such as clinical texts. These datasets often possess complex and unique structures, which require more specialized approaches. Notably, despite OpenPipe running two different LLM models under the hood, it still exhibited very poor performance on the clinical dataset — highlighting the limitations of generalized architectures in specialized, high-stakes domains. One likely reason for this performance discrepancy is that the general-purpose text used in this study may have been part of the data used during the fine-tuning of OpenPipe’s LLM models. In contrast, the clinical dataset falls outside that domain familiarity, making it more challenging for the models to perform effectively without targeted fine-tuning. This further emphasizes the shortcomings of general-purpose models in healthcare, where accuracy and reliability are non-negotiable.
In comparison, paid solutions, particularly those from John Snow Labs, demonstrated superior performance, especially in complex clinical texts. With significantly higher accuracy in both entity-level and token-level evaluations, commercial tools offer a more robust and reliable choice for organizations prioritizing regulatory compliance and precision in clinical text de-identification. Ultimately, the decision between open-source and commercial solutions depends on factors such as budget, expected performance, and the complexity of the use case. However, for high-stakes healthcare applications, commercial tools remain the most dependable option for achieving regulatory-grade de-identification.
For a more detailed analysis of our evaluation of commercial APIs and John Snow Labs’ performance on clinical data, you can refer to our academic paper “Can Zero-Shot Commercial APIs Deliver Regulatory-Grade Clinical Text De-Identification?” or check out the related blog post “Comparing Medical Text De-Identification Performance”.
You can find the whole code of this study in the Commercial_Solutions_VS_Open_Source_Models_for_Medical_Context notebook.
Beyond its standalone capabilities, John Snow Labs is also available on AWS, Snowflake, and Databricks Marketplaces, making it easier for organizations to integrate de-identification solutions into their existing workflows.
- Seamless Integration: Deploy directly within your preferred cloud ecosystem without a complex setup.
- Scalability: Process large-scale clinical data efficiently.
- Compliance & Security: Designed for healthcare-grade data protection, meeting HIPAA and GDPR standards.
Healthcare NLP models are licensed, so if you want to use these models, you can watch “Get a Free License For John Snow Labs NLP Libraries” video and request one license here.