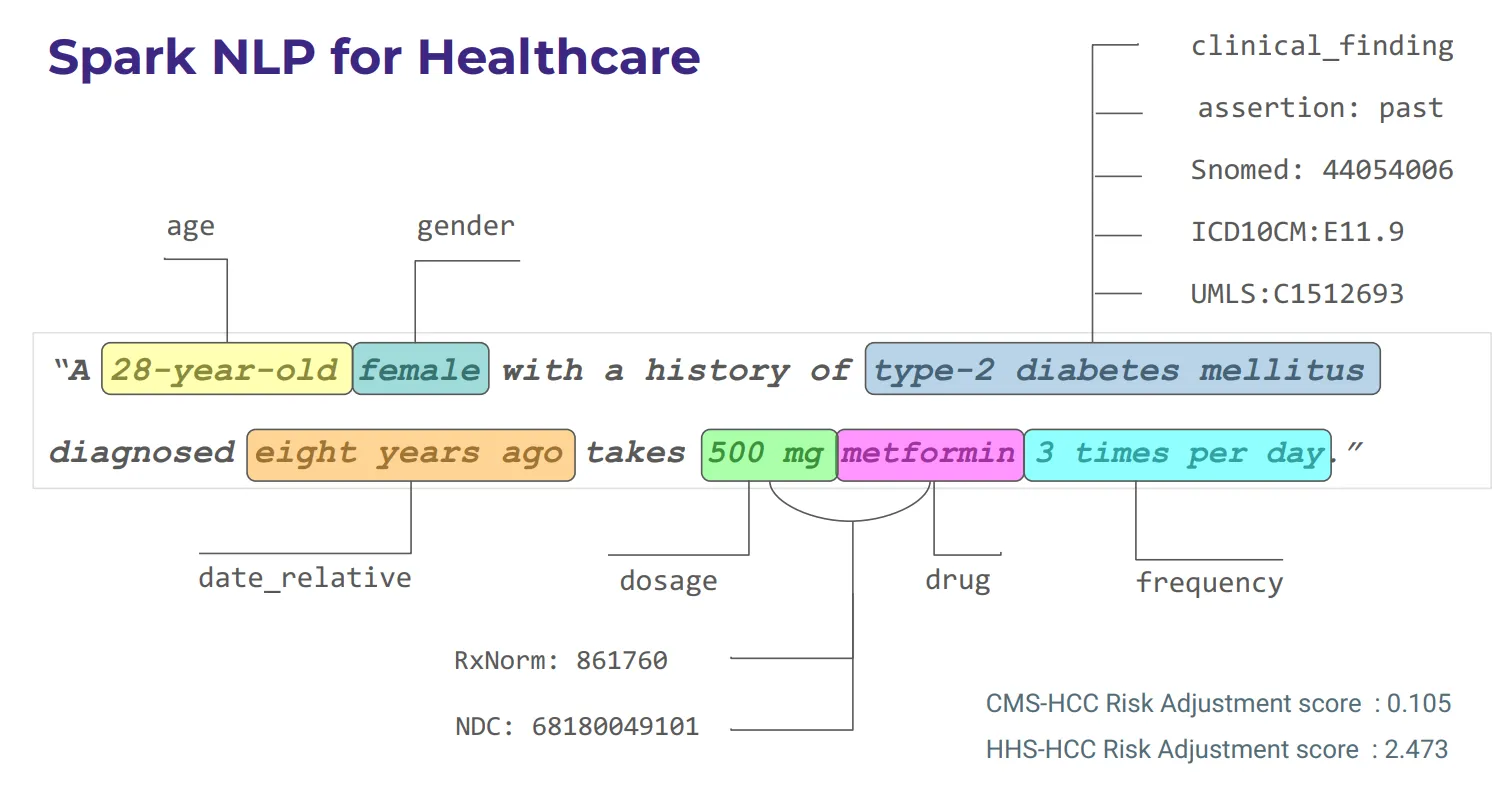
Healthcare-specific language models, like the JSL-MedS-NER family, are designed to extract clinical entities from unstructured medical text. These models can identify key information such as clinical terms, drugs, side effect, cancer diagnoses, metastasis, and protected health information (PHI). With multiple quantization options (q4, q8, q16), users can balance speed and accuracy for their specific needs, making these models highly effective for tasks like pharmacovigilance, oncology reporting, and clinical data processing
The ability to extract clinical entities from unstructured text — such as medical notes, clinical trial data, or patient records — has become increasingly critical in healthcare. Leveraging healthcare-specific large language models (LLMs) allows organizations to process vast amounts of medical data efficiently and with high accuracy. These LLMs are fine-tuned for the clinical and medical domains, enabling them to recognize complex medical entities that are relevant to patient health records, research, or treatment plans.
Healthcare NLP with John Snow Labs
The Healthcare NLP Library, part of John Snow Labs’ Library, is a comprehensive toolset designed for medical data processing. It includes over 2,400 pre-trained models and pipelines for tasks like clinical information extraction, named entity recognition (NER), and text analysis from unstructured sources such as electronic health records and clinical notes. The library is regularly updated and built on advanced algorithms to support healthcare professionals. John Snow Labs also offers a GitHub repository with open-source resources, certification training, and a demo page for interactive exploration of the library’s capabilities.
What is a Healthcare-Specific LLM?
Healthcare-specific LLMs are language models fine-tuned for the medical and clinical domains. These models are designed to process clinical text and recognize entities that are pertinent to patient health records, medical research, or other healthcare-related documents.
A key capability of such LLMs is their ability to extract entities relevant to PHI — such as names, locations, dates, contact details, and professions — while simultaneously identifying medical concepts, diagnoses, treatments, and symptoms.
The JSL-MedS-NER family of models offers powerful tools for extracting clinical entities from unstructured medical text. These models come with multiple quantization options (q4, q8, q16), allowing users to optimize performance based on their specific task requirements. Whether prioritizing speed, accuracy, or a balance of both, the JSL-MedS-NER models are designed to efficiently handle various healthcare-related entity extraction tasks, such as identifying drugs, adverse events, diagnoses, and patient information.
In this post, we’ll explore the use of these models for entity extraction, with a focus on the following areas: Posology and Adverse Drug Events, Oncology, Clinical Information, and PHI Deidentification.
-
Posology and Adverse Drug Event Extraction: Identifying Drugs and Side Effects
Understanding drug prescriptions (posology) and recognizing adverse drug events (ADE) is critical in clinical settings to ensure patient safety and treatment efficacy. John Snow Labs has achieved remarkable success with its Posology and ADE models, particularly in the field of named entity recognition (NER). You can explore the example Clinical Named Entity Recognition and Adverse Drug Event ADE NER and Classifier Notebooks to see how these models perform in action.
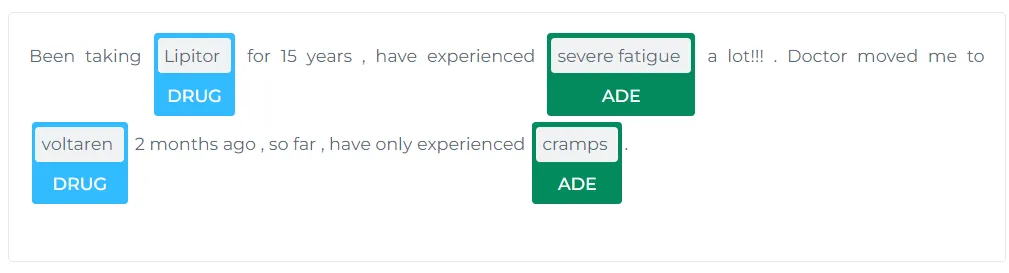
https://demo.johnsnowlabs.com/healthcare/ADE/
However, there may be instances where the desired entity cannot be extracted, or the predicted output may not fully meet the requirements, especially when applied to specialized domains such as medical and healthcare. In such cases, JSL-MedS-NER can serve as a valuable alternative, providing enhanced capabilities for recognizing complex medical entities that might be missed by other models. For instance, using JSL-MedS-NER, it’s possible to extract the following from patient records:
Example Input:
import json
text = """
I feel a bit drowsy & have a little blurred vision, and some gastric problems.
I've been on Arthrotec 50 for over 10 years on and off, only taking it when
I needed it.
"""
ents = {
"drugs": [],
"adverse drug event": []
}
ner_prompt = f"""<|input|>
### Template:
{json.dumps(ents, indent=4)}
### Text:
{text}
<|output|>
"""
Example Output:
{
"drugs": [
"Arthrotec"
],
"adverse drug event": [
"drowsy",
"blurred vision",
"gastric problems"
]
}
In the above example, the model accurately identifies “Arthrotec” as the drug and lists adverse events such as drowsiness, blurred vision, and gastric problems. These extractions assist in pharmacovigilance, supporting clinicians in managing medication risks effectively.
a. Extracting RxNorm Terminology Codes from Drug Entities Using John Snow Labs’ RxNorm Model
RxNorm is a standardized system for drug names and clinical drug information, maintained by the U.S. National Library of Medicine. Each medication is assigned a unique RxNorm code, which ensures consistent and accurate communication about drugs across healthcare systems. These codes are crucial for drug identification, prescription management, and pharmacovigilance.
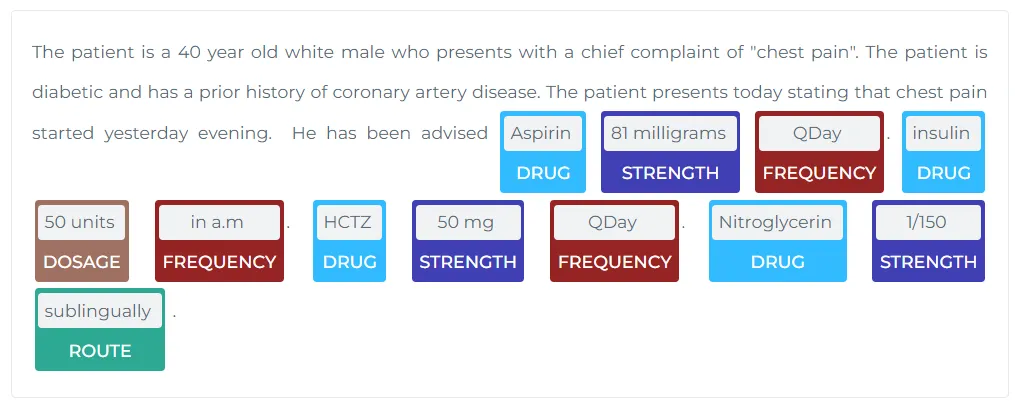
https://demo.johnsnowlabs.com/healthcare/NER_POSOLOGY/
In this task, drug names found in clinical text are extracted and mapped to their corresponding RxNorm codes using John Snow Labs’ Natural Language Processing (NLP) library. This approach uses the RxNorm model (sbiobertresolve_rxnorm_augmented) to connect the identified drugs to a standardized RxNorm database.
Example Input:
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("doc_ner_chunk")
sbert_embedder = BertSentenceEmbeddings\
.pretrained('sbiobert_base_cased_mli', 'en','clinical/models')\
.setInputCols("doc_ner_chunk")\
.setOutputCol("sentence_embeddings")\
.setCaseSensitive(False)
rxnorm_resolver = SentenceEntityResolverModel\
.pretrained("sbiobertresolve_rxnorm_augmented","en", "clinical/models")\
.setInputCols(["sentence_embeddings"]) \
.setOutputCol("rxnorm_code")\
.setDistanceFunction("EUCLIDEAN")
# Build up the pipeline
resolver_pipeline = Pipeline(
stages = [
document_assembler,
sbert_embedder,
rxnorm_resolver
])
Example Output:
| | chunk | code | resolutions | aux_labels |confidence | |--:|:----------|------:|:----------------------|:----------:|----------:| | 0 | Arthrotec | 68373 | arthrotec [arthrotec] | Brand Name | 0.995 |
The output shows that the drug “Arthrotec” is identified and mapped to its corresponding RxNorm code “68373” with a high confidence level of 0.995. This standardization ensures that “Arthrotec” can be recognized consistently across different healthcare platforms and systems.
This functionality, provided by the John Snow Labs library, simplifies the task of extracting drug entities from clinical text and translating them into universally recognized codes, facilitating improved healthcare interoperability.
b. Extracting ICD-10 Terminology Codes from Clinical Entities Using John Snow Labs’ ICD-10 Model
ICD-10 (International Classification of Diseases, 10th Revision) is a coding system used globally to classify diseases, symptoms, and medical conditions. Each condition or symptom has a unique ICD-10 code, which is vital for documenting clinical diagnoses, billing, and research.
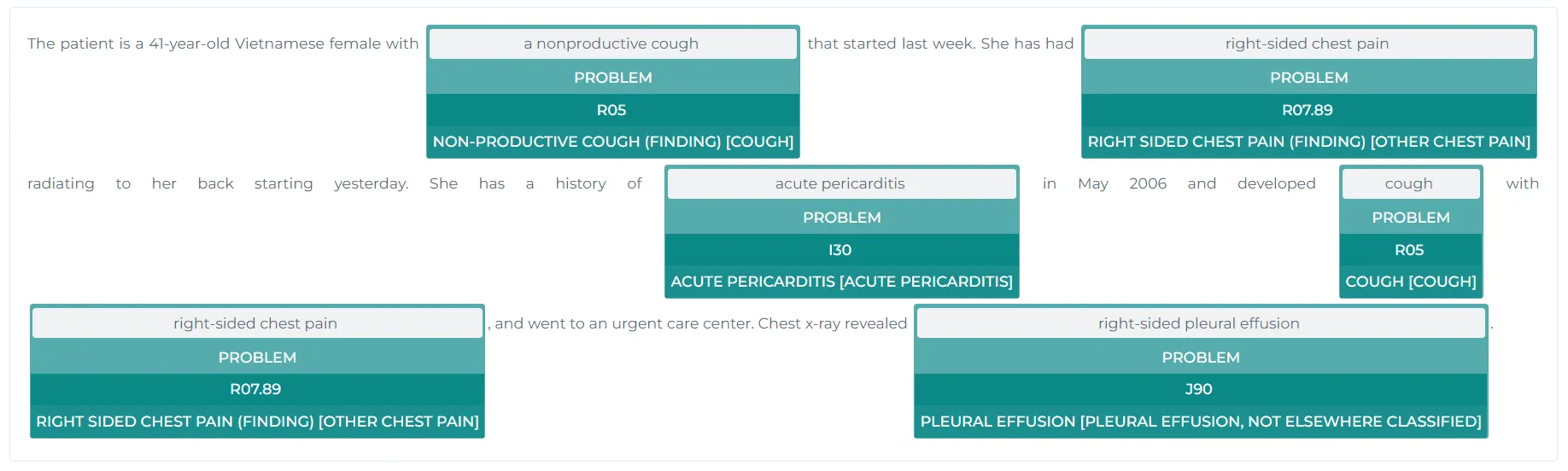
https://demo.johnsnowlabs.com/healthcare/ER_ICD10_CM/
This section demonstrates how to extract medical conditions (e.g., symptoms or diagnoses) from clinical text and map them to corresponding ICD-10 codes using John Snow Labs’ NLP library. The ICD-10 model (sbiobertresolve_icd10cm_augmented_billable_hcc) facilitates this process by standardizing these conditions to universally recognized ICD-10 codes. This is crucial for accurate diagnosis recording, billing, and understanding of the patient’s condition.
Example Input:
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("doc_ner_chunk")
sbert_embedder = BertSentenceEmbeddings\
.pretrained('sbiobert_base_cased_mli', 'en','clinical/models')\
.setInputCols("doc_ner_chunk")\
.setOutputCol("sentence_embeddings")
icd_resolver = SentenceEntityResolverModel\
.pretrained("sbiobertresolve_icd10cm_augmented_billable_hcc", "en", "clinical/models")\
.setInputCols(["sentence_embeddings"]) \
.setOutputCol("icd10cm_code")\
.setDistanceFunction("EUCLIDEAN")
# Build up the pipeline
resolver_pipeline = Pipeline(
stages = [
document_assembler,
sbert_embedder,
icd_resolver
])
Example Output:
| | chunk | code | resolutions |confidence| |--:|:-----------------|:------|:------------------------------------------------------------------------------|---------:| | 0 | drowsy | R40.0 | drowsy [somnolence] | 0.9926 | | 1 | blurred vision | H53.8 | blurred vision [other visual disturbances] | 0.9942 | | 2 | gastric problems | K31.9 | functional gastric disturbance [disease of stomach and duodenum, unspecified] | 0.2971 |
The output table shows how various clinical entities (e.g., “drowsy,” “blurred vision,” and “gastric problems”) are mapped to their respective ICD-10 codes:
- “Drowsy” is mapped to ICD-10 code “R40.0” (Somnolence) with a high confidence of 0.9926.
- “Blurred vision” is mapped to “H53.8” (Other visual disturbances) with a confidence of 0.9942.
- “Gastric problems” is mapped to “K31.9” (Functional gastric disturbance), though with lower confidence, indicating a less certain match.
By using John Snow Labs’ ICD-10 model, medical professionals can automatically extract and map conditions from clinical notes to ICD-10 codes, ensuring accurate and standardized documentation for diagnoses and medical billing.
These extractions (RxNorm and ICD-10) help improve clinical decision-making and communication across healthcare systems by leveraging standardized medical terminologies. For more terminology code examples such as RxNorm, ICD, SNOMED, CPT, UMLS, LOINC, and MeSH .etc please see the Clinical_Entity_Resolvers notebooks
-
Oncology, Cancer, Metastasis, and Stages
Cancer diagnosis, metastasis tracking, and treatment plans are often complex and involve a wide range of medical terminologies, procedures, and biomarkers. John Snow Labs has developed highly successful Oncology models, capable of extracting over 40 oncology-related entities, such as therapies, diagnostic tests, and staging information. These models provide robust solutions for many oncology use cases. You can explore the example Oncology Models Notebook to see how these models perform in action.
https://demo.johnsnowlabs.com/healthcare/ONCOLOGY/
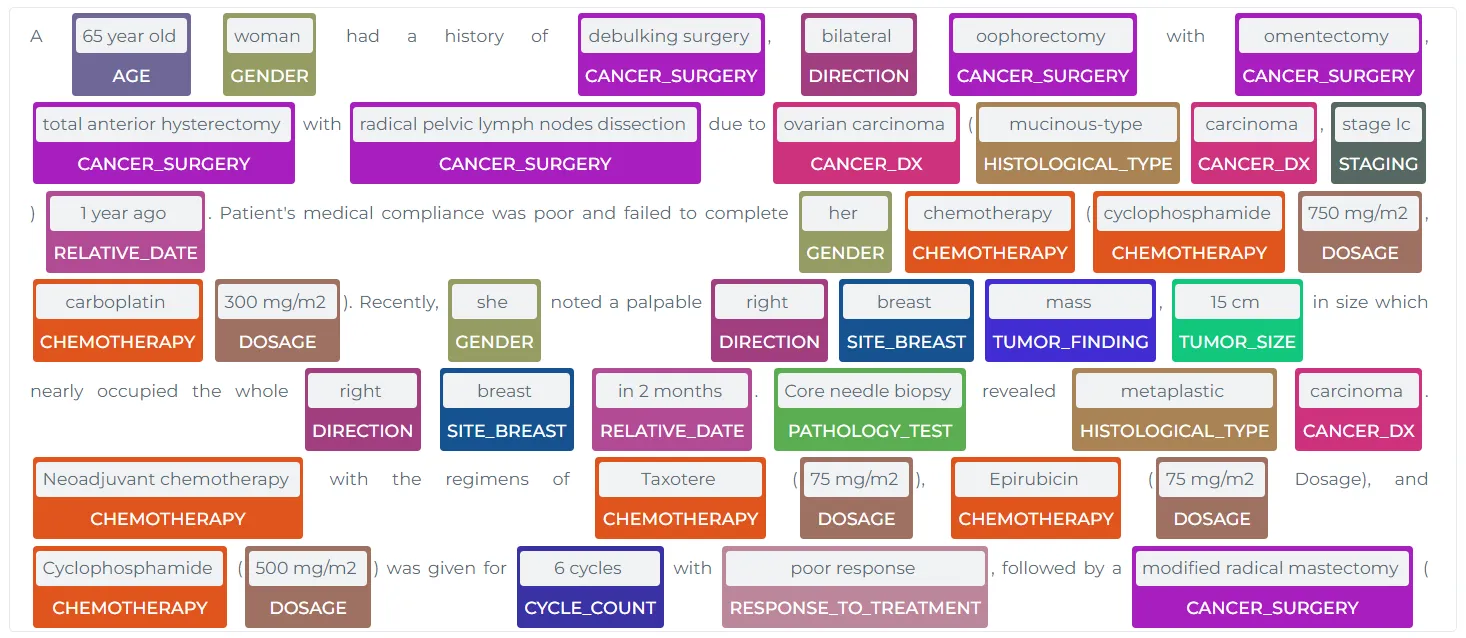
However, if a specific entity is not identified, or if the predicted entity from the NER models in the Models Hub is insufficient, users can turn to JSL-MedS-NER for more comprehensive entity recognition. Using models like JSL-MedS-NER, key clinical terms related to cancer, metastasis, tumor size, and chemotherapy regimens can be identified, as shown in the following example:
Example Input:
import json
text = """
A 65-year-old woman had a history of debulking surgery, bilateral oophorectomy
with omentectomy, total anterior hysterectomy with radical pelvic lymph nodes
dissection due to ovarian carcinoma (mucinous-type carcinoma, stage Ic) 1 year ago.
Patient's medical compliance was poor and failed to complete her chemotherapy
(cyclophosphamide 750 mg/m2, carboplatin 300 mg/m2). Recently, she noted a
palpable right breast mass, 15 cm in size which nearly occupied the whole right
breast in 2 months. Core needle biopsy revealed metaplastic carcinoma.
Neoadjuvant chemotherapy with the regimens of Taxotere (75 mg/m2), Epirubicin
(75 mg/m2 Dosage), and Cyclophosphamide (500 mg/m2) was given for 6 cycles with
poor response, followed by a modified radical mastectomy (MRM) with dissection
of axillary lymph nodes and skin grafting. Postoperatively, radiotherapy was
done with 5000 cGy in 25 fractions. The histopathologic examination revealed a
metaplastic carcinoma with squamous differentiation associated with adenomyoepithelioma.
Immunohistochemistry study showed that the tumor cells are positive for epithelial
markers-cytokeratin (AE1/AE3) stain, and myoepithelial markers, including
cytokeratin 5/6 (CK 5/6), p63, and S100 stains. The dissected axillary lymph
nodes showed metastastic carcinoma with negative hormone receptors in 3 nodes.
The patient was staged as pT3N1aM0, with histologic tumor grade III
"""
ents = {
"Biomarker": [],
"Cancer_Dx": [],
"Cancer_Surgery": [],
"Chemotherapy": [],
"Grade": [],
"Metastasis": [],
"Tumor_Size": [],
"Unspecific_Therapy": []
}
ner_prompt = f"""<|input|>
### Template:
{json.dumps(ents, indent=4)}
### Text:
{text}
<|output|>
"""
Example Output:
{
"Biomarker": [
"AE1/AE3",
"CK 5/6",
"p63",
"S100"
],
"Cancer_Dx": [
"ovarian carcinoma",
"metaplastic carcinoma",
"adenomyoepithelioma"
],
"Cancer_Surgery": [
"debulking surgery",
"bilateral oophorectomy",
"total anterior hysterectomy",
"modified radical mastectomy"
],
"Chemotherapy": [
"cyclophosphamide",
"carboplatin",
"Taxotere",
"Epirubicin"
],
"Grade": [
"III"
],
"Metastasis": [
"metastatic carcinoma"
],
"Tumor_Size": [
"15 cm"
]
}
Here, the model extracts relevant details such as tumor biomarkers, diagnosis (ovarian carcinoma, metaplastic carcinoma), and cancer treatment modalities, enabling precise documentation and research insights.
-
Clinical Information Extraction: Diagnoses, Treatments, Tests, and Symptoms
John Snow Labs offers highly successful Clinical Models designed for extracting critical clinical entities such as diagnoses, treatments, tests, symptoms, and more. These models excel in information extraction for various healthcare applications. You can explore the example Clinical Named Entity Recognition Notebook to see how these models perform in action.
https://demo.johnsnowlabs.com/healthcare/NER/
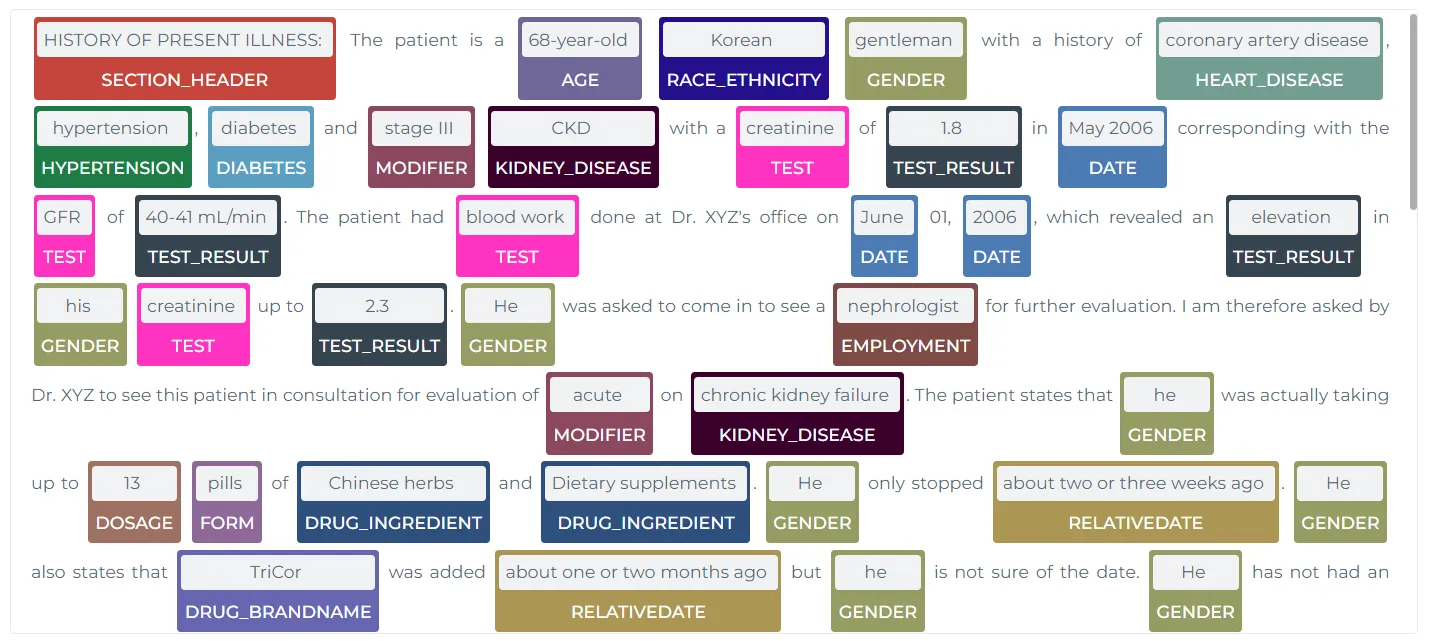
However, if a specific entity is not identified, or the predicted entity from the NER models in the Models Hub does not meet the requirement, users can rely on JSL-MedS-NER for enhanced entity recognition. These models can detect medical concepts such as diagnoses, treatments, symptoms, and test results, providing a comprehensive understanding of a patient’s condition. This capability supports clinical decision-making and documentation efforts, helping medical professionals to focus on critical aspects of patient care.
Example Output:
{
"Diagnosis": [
"hypertension",
"diabetes mellitus"
],
"Treatment": [
"insulin therapy",
"antihypertensive medication"
],
"Test Results": [
"elevated blood glucose",
"high blood pressure"
],
"Symptoms": [
"fatigue",
"headache"
]
}
This output highlights how the LLM can detect key medical concepts from a patient’s record, enabling clinicians to easily track diagnoses and treatment plans.
-
PHI Information and Deidentification: Protecting Patient Privacy
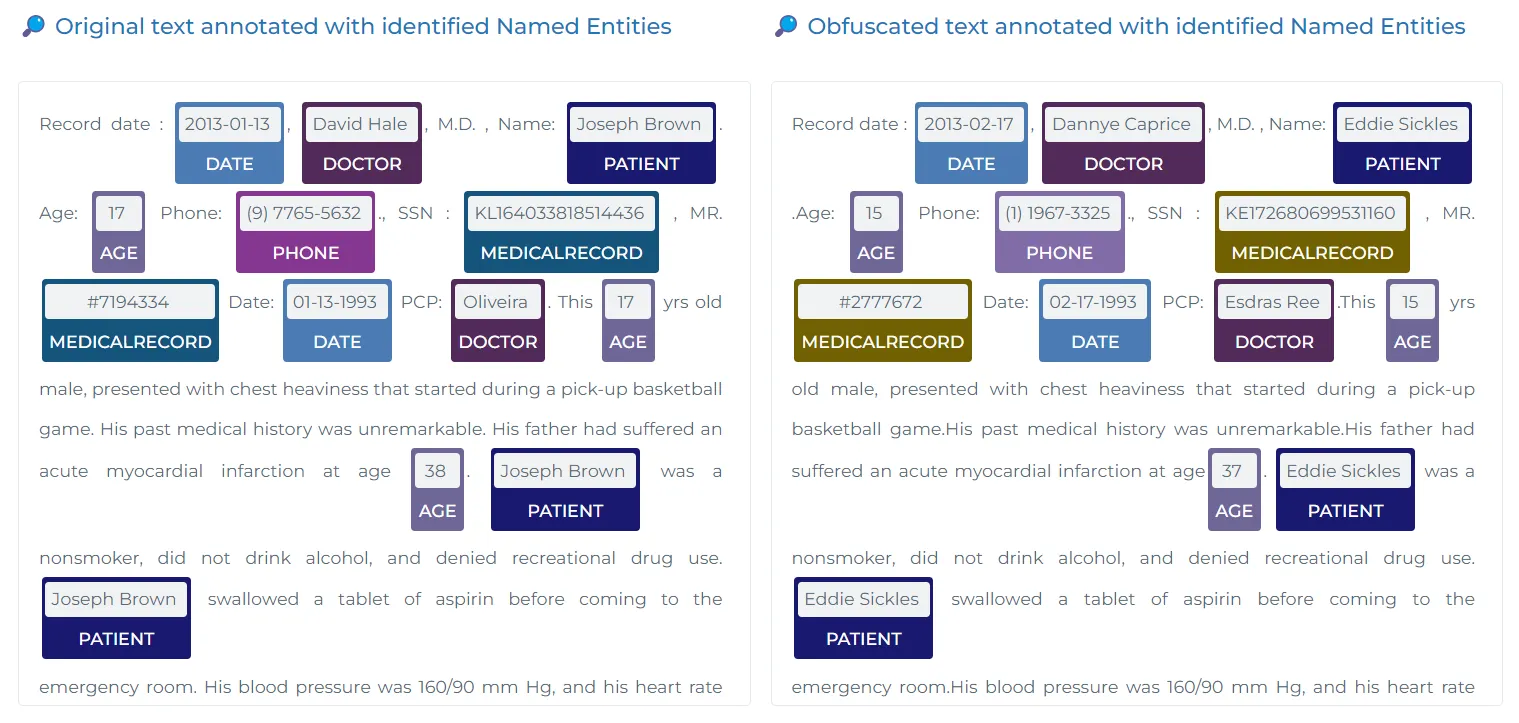
One of the critical aspects of healthcare data processing is ensuring the protection of personally identifiable information (PHI). John Snow Labs offers highly effective de-identification models that excel in extracting protected health information (PHI), including sensitive data such as Dates, Names, Locations, Professions, Contacts, Ages, IDs, and more. These models ensure robust PHI extraction for compliance and privacy purposes. You can explore the example PHI Entities and DeIdentification Notebook to see how these models perform in action.
https://demo.johnsnowlabs.com/healthcare/DEIDENTIFICATION_CONSISTENCY/
However, if a specific entity is not identified or if the predicted entity from the NER models in Models Hub is insufficient, users can turn to JSL-MedS-NER for more comprehensive entity recognition. These models help healthcare organizations comply with data privacy regulations like HIPAA.
Example Input:
import json
text = """
David Hale, M.D. he is 45 years old and works as Data Engineer at GE
Healthcare, which is located in Delaware. For personal Contact and
Email: 555-555-555, hale@gmail.com, Dr. Molina Cortes, MD. is
affiliated with the Cocke County Baptist Hospital in Des Moines.
"""
ents = {
"DATE": [],
"NAME": [],
"AGE": [],
"LOCATION": [],
"PROFESSION": [],
"CONTACT": [],
"Email": []
}
ner_prompt = f"""<|input|>
### Template:
{json.dumps(ents, indent=4)}
### Text:
{text}
<|output|>
"""
Example Output:
{
"DATE": [],
"NAME": [
"David Hale",
"Molina Cortes"
],
"AGE": [
"45"
],
"LOCATION": [
"Delaware",
"Cocke County",
"Des Moines"
],
"PROFESSION": [
"Data Engineer"
],
"CONTACT": [
"555-555-555"
],
"Email": [
"hale@gmail.com"
]
}
In this case, the LLM accurately identifies sensitive PHI such as names, contact details, and locations, allowing the data to be safely anonymized for research or compliance purposes
Choosing the Right Quantization for Your Use Case
Both JSL-MedS-NER-v1 and JSL-MedS-NER-v2 come in different quantization options (q4, q8, q16), offering varying levels of computational efficiency and accuracy.
- q4: Offers the fastest processing speed, ideal for large-scale data processing with moderate accuracy needs.
- q8: Provides a balance between accuracy and speed, making it suitable for most clinical entity extraction tasks.
- q16: Maximizes accuracy, recommended for critical tasks requiring high precision, such as oncology reports or pharmacovigilance.
Advantages of Using Healthcare-Specific LLMs for Clinical Entity Extraction
- Improved Accuracy in Medical Data Processing
Healthcare-specific LLMs are fine-tuned for medical language, ensuring more accurate identification of drugs, diagnoses, and treatments compared to general-purpose models. - Efficient Handling of Large Volumes of Data
With quantization options (q4, q8, q16), these models can be optimized for faster processing, allowing healthcare providers to manage vast amounts of unstructured data efficiently. With multiple quantization levels, users can tailor the model’s speed and accuracy to their specific needs, whether it’s rapid processing of records or detailed analysis of complex cases. - Versatile Application Across Clinical Domains
From oncology reports to routine medical notes, these models can extract key clinical concepts, enabling insights across a wide range of medical specialties. By accurately identifying adverse drug events and interactions, these models support better pharmacovigilance and reduce the risks associated with medication errors. - Compliance with Data Privacy Regulations
The ability to deidentify sensitive patient information (PHI) ensures compliance with data protection laws like HIPAA, making it safer to use clinical data for research or analytics.
For additional information, please see the following references:
- MedicalLLM and LLMLoader’s pretrained models: Loading Medical and Open-Source LLMs
- Various RAG Implementations with JohnSnowLabs: generative-ai
- Clinical Entity Resolvers Models (Terminology Code Examples)
- Clinical Named Entity Recognition
- Adverse Drug Event ADE NER and Classifier
- Oncology Models
- PHI Entities and DeIdentification
- Generative AI in Healthcare
Conclusion
Healthcare-specific LLMs, such as the JSL-MedS-NER family, are revolutionizing the extraction of clinical entities from unstructured medical data. These models possess an advanced ability to accurately identify and interpret complex medical terminologies, treatments, adverse events, and other critical patient information, which is often difficult to process through traditional methods. By automating these tasks, healthcare professionals can focus on delivering care with greater precision and efficiency.
Moreover, these models not only enhance productivity but also play a crucial role in maintaining the integrity and confidentiality of patient data, as they are designed to meet stringent standards for data privacy and security in the healthcare domain. As these models continue to evolve, they hold immense potential to improve clinical decision-making, support research efforts, and pave the way for more personalized and data-driven healthcare practices, ultimately improving patient outcomes across diverse medical settings.