This blog post explores how John Snow Labs’ Healthcare NLP and LLM library is transforming cancer care by enhancing the interpretation of medical records. Focusing on six common cancer types, the article demonstrates how advanced natural language processing is improving diagnosis, prognosis, and personalized treatment planning by extracting and analyzing critical information from unstructured clinical data.
Cancer is one of the leading causes of death worldwide, with millions of new cases diagnosed each year. In 2020 alone, over 19 million new cancer cases were reported globally, with this number expected to rise due to population growth and aging. The complexity of cancer makes treatment challenging, as it is not a single disease but a group of related conditions that can affect virtually any part of the body.
The future of global cancer rates paints a concerning picture. A recent projection below shows new cancer cases worldwide are expected to surge from about 14 million in 2012 to nearly 24 million by 2035. Alongside this, cancer deaths are predicted to rise from around 8 million to 14.5 million in the same period. These stark numbers highlight the growing urgency of cancer research, prevention, and treatment on a global scale.
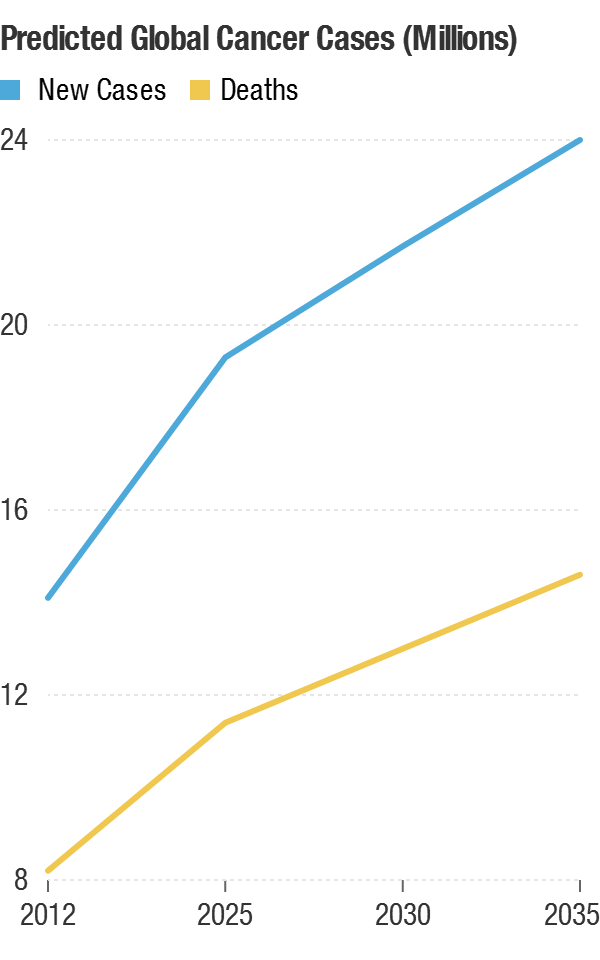
Source: World Health Organization; Credit: Michaeleen Doucleff/NPR
By understanding the underlying mechanisms of cancer, developing early detection methods, and personalizing treatment approaches, researchers aim to improve outcomes and reduce the burden of this disease. However, the vast amount of clinical data generated during diagnosis, treatment, and follow-up care presents its own challenges. Extracting valuable information from unstructured clinical texts, such as identifying specific cancer types, is vital for research, patient care, and advancing the fight against cancer. This is where advancements in Natural Language Processing (NLP), particularly Named Entity Recognition (NER) models, can play a crucial role.
In the context of cancer research, NER models can identify and categorize mentions of cancer types, treatments, genes, and other medically relevant entities in clinical texts, such as patient records, pathology reports, and research articles. By doing so, they help convert vast amounts of text into structured data that can be easily analyzed and utilized for research and clinical decision-making.
John Snow Labs, renowned for its innovative contributions to healthcare data science, provides a comprehensive NLP library specifically designed to address the challenges in the analysis of cancers in medical records. This library leverages state-of-the-art techniques, including NER, assertion and relation extraction models to extract meaningful insights from unstructured text. By harnessing these tools, healthcare professionals can uncover critical information on cancer, enabling more precise diagnosis, treatment, and prevention strategies.
This post covers using the brand new ner_cancer_types model in the Healthcare NLP library from John Snow Labs to extract entities related to cancer types.
Let us start with a short Spark NLP introduction and then discuss the details of analysis of types with some solid results.
Healthcare NLP & LLM For Interpretation Data About Cancers in Medical Records
The Healthcare Library is a powerful component of John Snow Labs’ Spark NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,200 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs has created custom large language models (LLMs) tailored for diverse healthcare use cases. These models come in different sizes and quantization levels, designed to handle tasks such as summarizing medical notes, answering questions, performing retrieval-augmented generation (RAG), named entity recognition and facilitating healthcare-related chats.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Spark NLP and related tools.
John Snow Labs also offers periodic certification training to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
Recognizing the Most Common Cancer Types from Clinical Notes
This NER model is designed to extract key cancer-related information from clinical and biomedical text, focusing on six primary cancer types. These categories represent a broad range of cancers, from those affecting the central nervous system to those originating in epithelial or connective tissues. In addition to identifying cancer types, the model extracts crucial context, such as the presence of metastasis, which indicates cancer spread, and biomarkers, including both their measurable quantities and results. By identifying these entities (shown below), the model helps clinicians and researchers obtain detailed insights into cancer diagnosis, treatment planning, and prognosis from unstructured clinical texts.

Entities extracted by ner_cancer_types_wip
Spark NLP uses pipelines and for extracting valuable information, only 6 stages will be needed.
# Step 1: Transforms raw text to `document`type
document_assembler = DocumentAssembler() \
.setInputCol("text") \
.setOutputCol( "document")
# Step 2: Sentence Detection/Splitting
sentence_detector = SentenceDetectorDLModel.pretrained("sentence_detector_dl_healthcare","en","clinical/models") \
.setInputCols(["document"]) \
.setOutputCol("sentence")
# Step 3: Tokenization
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")\
# Step 4: Clinical Embeddings
word_embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical", "en", "clinical/models") \
.setInputCols(["sentence", "token"]) \
.setOutputCol("embeddings")
# NER Model
ner_cancer_types = MedicalNerModel.pretrained("ner_cancer_types_wip", "en", "clinical/models") \
.setInputCols(["sentence","token", "embeddings"]) \
.setOutputCol("cancer_types_ner")
ner_cancer_types_converter = NerConverterInternal() \
.setInputCols(["sentence","token","cancer_types_ner"]) \
.setOutputCol("cancer_types_chunk")\
# Define the pipeline
nlp_pipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
word_embeddings,
ner_cancer_types,
ner_cancer_types_converter,
])
# Create an empty dataframe
data = spark.createDataFrame([[""]]).toDF("text")
# Fit the dataframe to the pipeline to get the model
nlp_model = nlp_pipeline.fit(data)
Let’s evaluate the model’s performance using this sample clinical text.
text = """ A 56-year-old man presented with a 2-month history of whole-body weakness, double vision, difficulty swallowing, and a 45 mm anterior mediastinal mass detected via chest CT. Neurological examination and electromyography confirmed a diagnosis of Lambert-Eaton Myasthenic Syndrome (LEMS), associated with anti-P/Q-type VGCC antibodies. A video-assisted thoracic surgery revealed histopathological features consistent with small cell lung cancer (SCLC) with lymph node metastases. The immunohistochemical analysis showed positive markers for AE1/AE3, TTF-1, chromogranin A, and synaptophysin. Notably, a pulmonary nodule in the left upper lobe disappeared, and FDG-PET/CT post-surgery revealed no primary lesions or metastases. """
Next stage is converting the unstructured text into a structured format. Structuring entities into a standardized format enhances data usability and integration, enabling efficient retrieval and comprehensive analysis of patient information. This process improves consistency and standardization, supporting advanced analytics and enabling better decision-making. Ultimately, it converts raw data into actionable insights, driving improvements in patient care, research outcomes, and public health strategies.
PipelineTracer is a recent addition to Healthcare NLP Library, that monitors every step of a pipeline, offering detailed information on entities, assertions, de-identification, classification, and relationships. It also aids in building parser dictionaries for creating a PipelineOutputParser. Key features include printing pipeline schemas, generating parser dictionaries, and retrieving potential assertions, relationships, and entities. It provides easy access to parser dictionaries and existing pipeline diagrams.
Let’s use the PipelineOutputParser to get the results as a dictionary:
from sparknlp_jsl.pipeline_output_parser import PipelineOutputParser
light_model = LightPipeline(nlp_model)
light_result = light_model5.fullAnnotate(text)
column_maps = {
'document_identifier': 'ner_cancer_types_wip',
'document_text': 'document',
'entities': ['cancer_types_chunk'],
'assertions': [],
'resolutions': [],
'relations': [],
'summaries': [],
'deidentifications': [],
'classifications': []
}
pipeline_parser = PipelineOutputParser(column_maps)
result = pipeline_parser.run(light_result)
result['result'][0]
PipelineOutputParser provides the results according to the predefined columns:
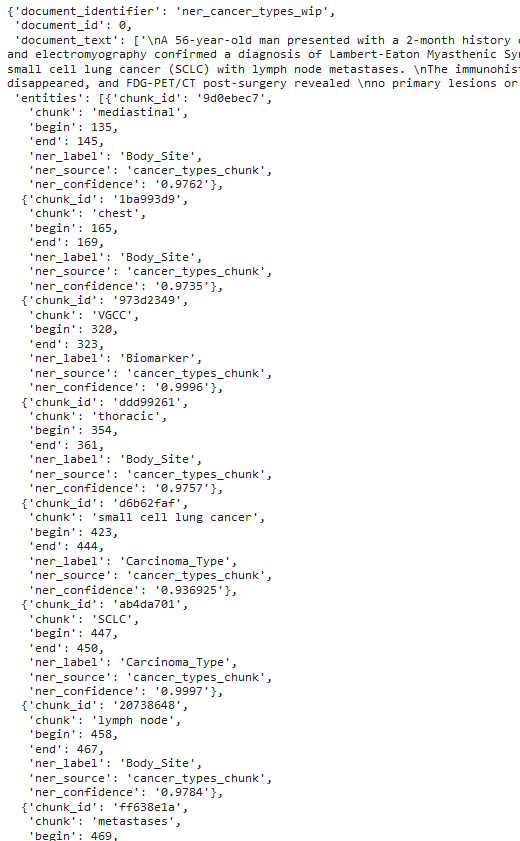
Detailed information extracted from the text (first 8 extracted entities are shown).
It is also possible to get the labelled entities as a Pandas dataframe and download the results:
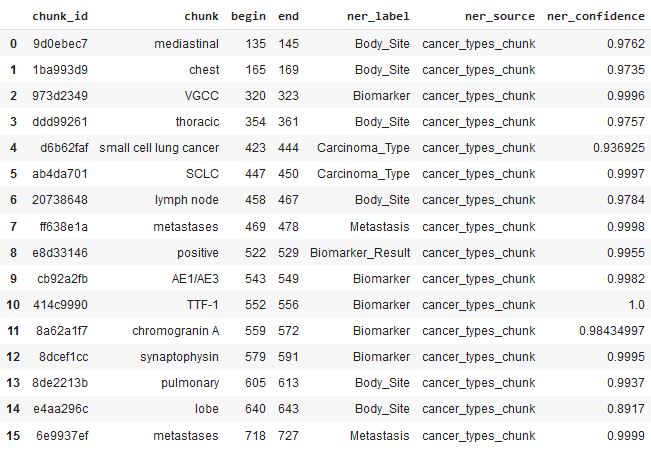
Dataframe of the Extracted Entities
John Snow Labs’ NER Visualizer offers a user-friendly interface for visualizing NER model outputs by highlighting and categorizing the entities detected within the text. This tool enables users to see how the models extract and label entities, making it easier to interpret and understand the data. The visualizer aids in validating model accuracy, identifying patterns, and deriving insights from unstructured medical data, ultimately supporting improved data analysis and decision-making in healthcare.
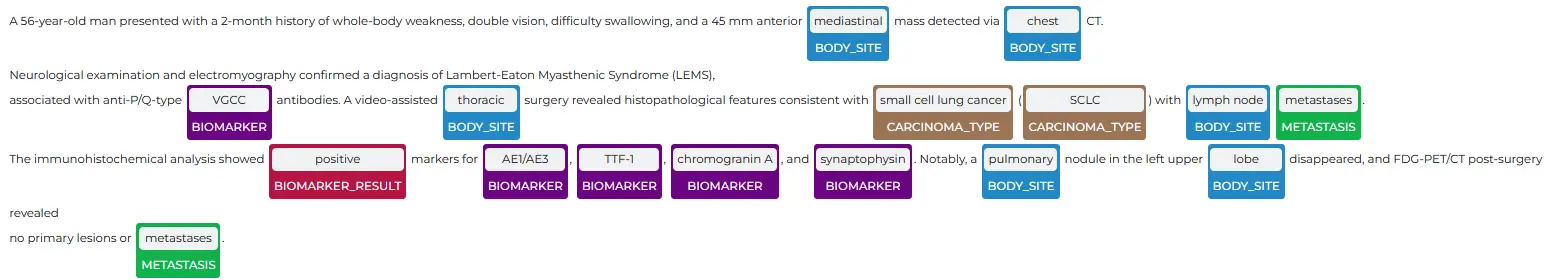
The NER model demonstrated a strong ability to identify cancer types and crucial cancer-related details from the sample clinical report.
Conclusion
John Snow Labs’ Healthcare NLP and LLM library is playing a transformative role in the way medical records are leveraged to enhance cancer care. By focusing on six of the most common cancer types, this advanced NER model enables healthcare professionals to extract, categorize, and analyze vital information from unstructured clinical data with greater precision.
This capability is not only improving the accuracy of diagnoses but also contributing to better prognoses and more tailored treatment plans for individual patients. With the ability to identify critical patterns and trends during the research of cancers in medical records, these tools are empowering clinicians to make more informed decisions, driving advancements in both patient care and cancer research. As NLP technology continues to evolve, its potential to further impact cancer treatment and public health strategies will be even more significant, helping to shape the future of oncology.





























