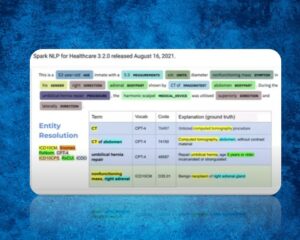
With the effect of digitalization, more and more data is collected in the field of health, as in every field. Using Spark NLP for Healthcare models, we can now derive very important inferences from irregular clinical notes. However, clinical terms in irregular clinical notes may not always be consistent with those in official sources. So, it is now possible to use Sentence Entity Resolver Models trained using official terms by adding new terms or modifying existing terms with Spark NLP for Healthcare v3.2!
Users can simply provide the path to any existing pretrained Sentence Entity Resolver model and train it further on the new dataset:
finetuned_model = bertExtractor.setPretrainedModelPath("existing_pretrained_model").fit(new_data)
Existing Sentence Entity Resolve Model
Let’s assume that we trained a SNOMED entity resolver model and saved this model to use later on.
bertExtractor = SentenceEntityResolverApproach()\
.setNeighbours(25)\
.setThreshold(1000)\
.setInputCols("bert_embeddings")\
.setNormalizedCol("_term")\
.setLabelCol("conceptId")\
.setOutputCol('snomed_code')\
.setDistanceFunction("EUCLIDIAN")\
.setCaseSensitive(False)
snomed_model = bertExtractor.fit(snomed_data)
# save if you will need that later
snomed_model.write().overwrite().save("sbiobertresolve_snomed_model")
Use Cases
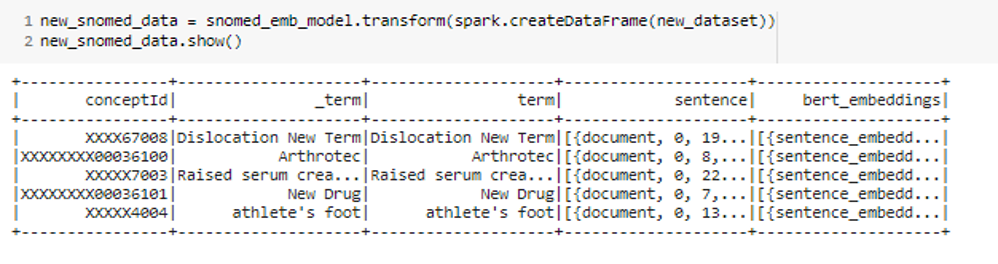
We can fine-tune this SNOMED model with a new dataset. In this case, we wanted to show you some use cases where you can see the success of the retraining feature, and we used `XXXX` in the codes to make it easier to see the difference easily.
- Added new rows that are close to their version in the main dataset (`Dislocation of joint -> Dislocation New Term`).
- Changed the code of terms existing in the main dataset (`Arthrotec`)
- Changed the positions of the words in the terms (`Serum creatinine raised -> Raised serum creatinine`)
- Added new terms that are not in the dataset (`New Drug`, `athlete’s foot`)
Adding Embedding Column
We need an embeddings column for training/ fine-tuning a sentence entity resolver model, so add that to our new dataframe.
documentAssembler = DocumentAssembler()\
.setInputCol("_term")\
.setOutputCol("sentence")
bert_embeddings = BertSentenceEmbeddings.pretrained("sbiobert_base_cased_mli", "en", "clinical/models")\
.setInputCols(["sentence"])\
.setOutputCol("bert_embeddings")
snomed_emb_model = PipelineModel(stages = [
documentAssembler,
bert_embeddings])
Fine-tuning Sentence Entity Resolver Model
We can simply provide the path to our pretrained Sentence Entity Resolver model by using new `setPretrainedModelPath` parameter and train it further on the new dataset.
new_snomed_model = bertExtractor.setPretrainedModelPath("sbiobertresolve_snomed_model").fit(new_snomed_data)
Test New Model Results
We will create a pipeline that contains both of the models and check the results by using `LightPipeline`.
documentAssembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("ner_chunk")
sbert_embedder = BertSentenceEmbeddings.pretrained('sbiobert_base_cased_mli', 'en','clinical/models')\
.setInputCols(["ner_chunk"])\
.setOutputCol("sbert_embeddings")
first_model = SentenceEntityResolverModel.load("sbiobertresolve_snomed_model") \
.setInputCols(["ner_chunk", "sbert_embeddings"]) \
.setOutputCol("first_code")
second_model = SentenceEntityResolverModel.load("new_sbiobertresolve_snomed_model") \
.setInputCols(["ner_chunk", "sbert_embeddings"]) \
.setOutputCol("second_code")
pipelineModel = PipelineModel(
stages = [
documentAssembler,
sbert_embedder,
first_model,
second_model])
snomed_lp = LightPipeline(pipelineModel)
Let’s test our models with the terms that we added into the new dataset and the ones that come from the main model (`Stomach ache`).
chunk_list = ["Dislocation New Term", "Arthrotec", "Raised serum creatinine", "New Drug", "athlete's foot", "Stomach ache"]

Conclusion
As you can see in the results:
- The resolutions of the new added terms are at the top of the results
- The terms that we changed the concept_codes, are at the top of the results
- The close terms results are resolved successfully
- The terms that comes from the main dataset are resolved with the same result