This post explores how John Snow Labs’ Healthcare NLP & LLM library can be used to extract genes and phenotypes from clinical text. By leveraging NLP techniques, we can transform unstructured medical data into actionable insights, enabling more efficient genetic research, clinical diagnostics, and personalized medicine. The blog covers the key steps in training NER and assertion status detection models for this task, including data preparation, annotation, and evaluation, and highlights real-world use cases where extracting genetic and phenotypic entities enhances precision in clinical decision-making.
Introduction
In modern medicine, the unstructured data buried within clinical documents holds immense potential for breakthroughs in genetics and personalized healthcare. As genomic research continues to grow and finds its way into clinical practice, the need to extract meaningful insights from text-based medical records has never been greater. Whether it’s identifying gene mutations linked to disease risk or capturing phenotypic traits associated with genetic disorders, processing this information efficiently is key to advancing precision medicine. Yet, the sheer volume and complexity of these texts remain a significant challenge.
Named Entity Recognition (NER) is a powerful Natural Language Processing (NLP) technique for identifying and categorizing specific entities within unstructured text. By training an NER model tailored specifically to genetics and phenotypes, we can uncover valuable insights hidden in clinical documentation. This approach allows for the automated extraction of genetic markers, phenotypic descriptions, and their related details, transforming unstructured data into actionable knowledge.
This blog post discusses the NER model (ner_genes_phenotypes) that excels at identifying critical genetic information and phenotypic traits from clinical texts. Also, the assertion status detection model, assertion_genomic_abnormality_wip model classifies entities into distinct assertion categories, adding critical context to the extracted information. By automating the extraction of these complex entities, researchers and clinicians can save valuable time, reduce errors, and focus on making data-driven decisions that ultimately improve patient outcomes.
Through real-world examples and practical insights, this article highlights the transformative potential of applying John Snow Labs’ related Healthcare NLP models to the field of genetics and phenotypes. This post will demonstrate how those models can be the crucial to extract, organize, and utilize genetic and phenotypic data at scale.
John Snow Labs, offers a powerful NLP & LLM library tailored for healthcare, empowering professionals to extract actionable insights from medical text. Utilizing advanced AI techniques like NER, assertion status detection, relation extraction, Question-Answering, and summarizing, this library helps uncover vital genetics information for more accurate diagnosis, treatment, and prevention.
Let us start with a short Healthcare NLP introduction and then discuss the applications of John Snow Labs’ Healthcare NLP & LLM library in genetics.
Healthcare NLP
The Healthcare Library is a powerful component of John Snow Labs’ Healthcare NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,500 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs has created custom large language models (LLMs) tailored for diverse healthcare use cases. These models come in different sizes and quantization levels, designed to handle tasks such as summarizing medical notes, answering questions, performing retrieval-augmented generation (RAG), named entity recognition and facilitating healthcare-related chats.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Healthcare NLP and related tools.
John Snow Labs also offers periodic certification training to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
The Importance of Genetic Information and Related Diseases
Genetic information is the cornerstone of modern healthcare and biomedical research, unlocking the secrets of our DNA to improve diagnosis, treatment, and prevention of diseases. With the rapid growth of genomic data, understanding how genetic variations and mutations influence health is crucial for advancing precision medicine. Diseases influenced by genetic factors, from rare disorders to complex conditions like cancer, rely on accurate identification and interpretation of genetic and phenotypic markers.
The dynamic relationship between genes and their functions, and how they express in various clinical and phenotypic contexts is key to developing effective diagnostic tests and personalized treatments. This knowledge enables healthcare providers to identify hereditary conditions, predict disease susceptibility, and tailor interventions that target specific genetic mutations.
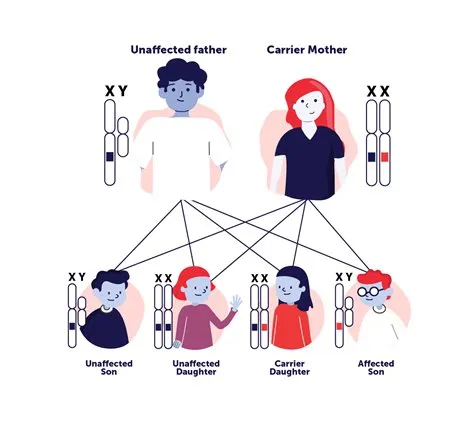
https://patienteducation.asgct.org/patient-journey/identifying-a-genetic-disease
NER models are essential for identifying and categorizing entities within text, such as genes, phenotypes, mutations, and clinical conditions. In the context of genetic and phenotypic analysis, ner_genes_phenotypes can automatically recognize mentions of genes, phenotypic diseases, mutations, diagnostic tests, inheritance patterns, and other key entities (a total of 17 entities) from large volumes of unstructured clinical or biomedical documentation.
Here are the 17 entities extracted by this model:
Clinical_Presentation: Medical signs and symptoms of a condition.
Diagnostic_Test: Medical procedures used to identify or assess a genetic disease.
Gene: Specific genetic code or sequence, including variants like F508del.
Gene_Diversity: Genetic variation within a population.
Gene_Function: Role or purpose of a specific gene or allele.
Gene_Interaction: How genes and environment interact to influence trait expression.
Gene_Penetrance: Percentage of individuals with a genetic mutation who show the associated trait.
Incidence: Number of new disease cases in a population over a specific time period.
Inheritance_Pattern: How genetic traits are passed between generations.
MPG: Names and abbreviations of Molecules, Proteins, and Genes.
OMIM: Comprehensive database of human genes and genetic disorders.
Other_Disease: Non-genetic diseases like infections or injuries.
Phenotype_Disease: Specific disease manifestations influenced by genetic causes.
Prevalence: Population groups affected by a disease.
Site: Location of a genetic mutation on the genome.
Treatment: Medical strategies to manage or improve genetic disorders.
Type_Of_Mutation: Specific type of genetic change or variation.
Extracting Entities from Clinical/Medical Text
By systematically extracting and organizing this information, NER models create structured datasets that serve as the basis for advanced analysis.
Please check this notebook; the results for the first gene (ATP7B) are shown below.
Converting entities into a structured format enhances usability and integration, facilitating efficient retrieval and in-depth analysis of patient data. This approach improves consistency and standardization, enabling sophisticated analytical methods and more informed decision-making. Ultimately, it transforms unstructured data into actionable insights, driving better patient care, more effective research, and informed public health initiatives.
In the example below, the text covers the Wilson’s disease, which is a rare genetic disorder that causes copper to build up in the body, particularly in the liver, brain, and eyes. The cause is a mutation in the ATP7B gene, which is responsible for removing excess copper from the body.
The NER model clearly identified and labelled many entities, including the related Gene (ATP7B), the phenotype disease (Wilson’s disease), clinical presentations (symptoms) and more entities.
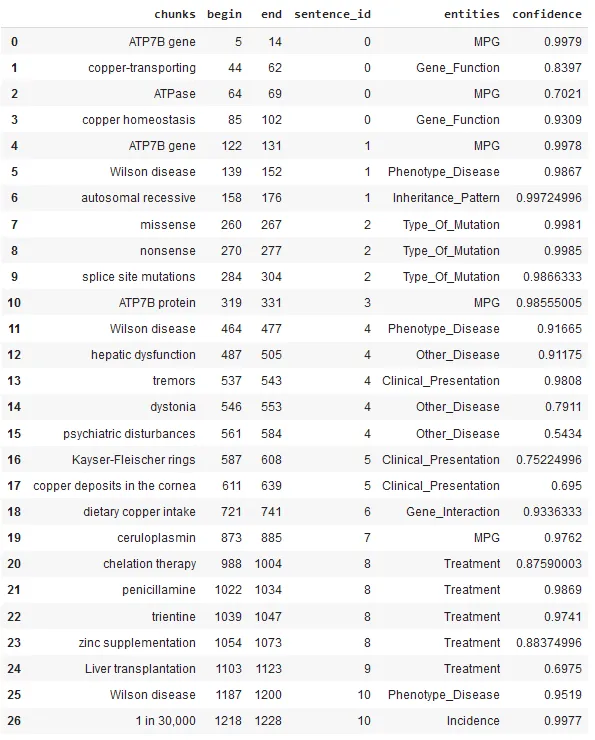
Extracted data in structured format.
John Snow Labs’ NER Visualizer offers an intuitive interface for exploring the outputs of NER models. It highlights and categorizes identified entities directly within the text, allowing users to clearly see how the models extract and label information.
This tool simplifies the process of understanding and interpreting extracted data, aiding in model validation, pattern recognition, and insight generation from unstructured medical text. By enhancing data analysis and interpretation, the visualizer supports more informed decision-making in healthcare.
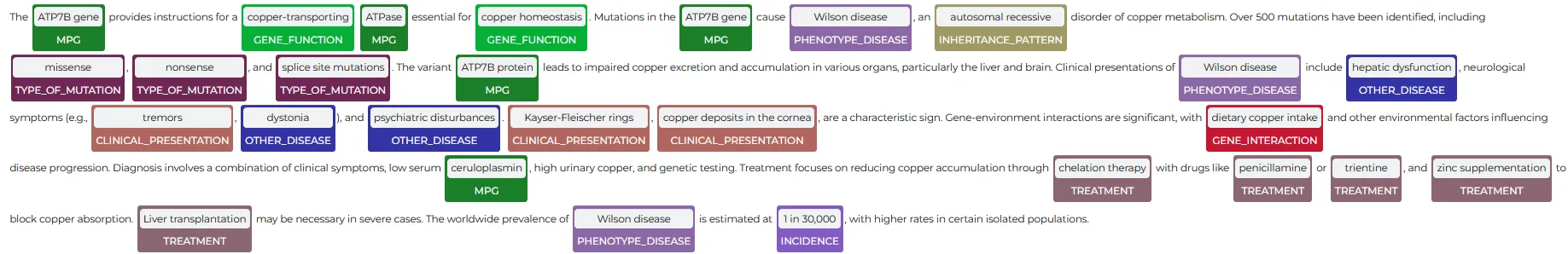
The NerVisualizer highlights the named entities that are identified by ner_genes_phenotypes and also displays their labels as decorations on top of the analyzed text.
Assertion Status Detection
The assertion_genomic_abnormality_wip model is designed to classify entities extracted by the ner_genes_phenotypes NER model into distinct assertion categories, adding critical context to the extracted information. It focuses on classifying entities such as Gene and MPG (Molecule-Protein-Gene) into one of three categories:
Normal: Indicates genes and molecules functioning as part of standard physiological processes without any abnormalities or mutations.
Affected: Identifies molecules or proteins impacted by genetic mutations, associating them with pathological or altered states.
Variant: Labels genes that are abnormal or represent a specific variant type, emphasizing genomic variations crucial for understanding genetic disorders or individual differences.
The assertion model adds precision to genomic and molecular analyses, enabling researchers and healthcare professionals to better interpret genetic data. This enriched context supports more accurate diagnostics, personalized treatment strategies, and advancements in genomics-driven healthcare.
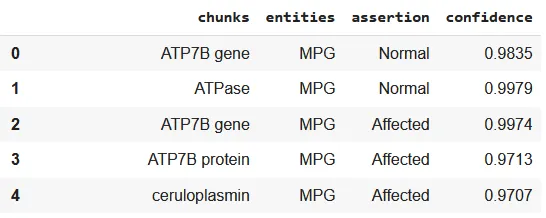
Assertion labels in structured format.
The Assertion Visualizer is a special type of NerVisualizer that also displays on top of the labeled entities the assertion status that was inferred by a Healthcare NLP model.
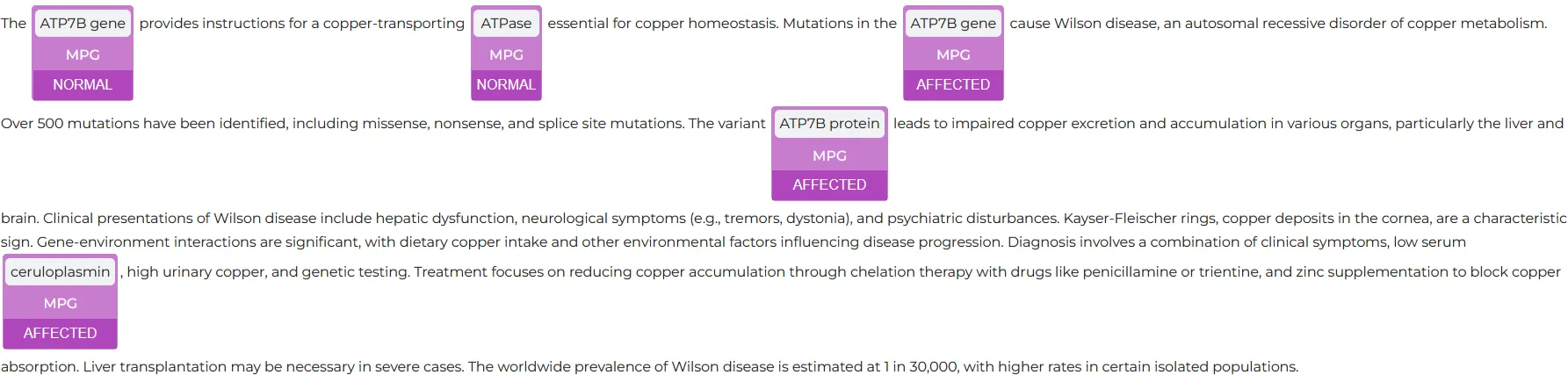
The AssertionVisualizer not only exhibits labeled entities but also overlays the assertion status of entities inferred by the NER model.
Conclusion
The integration of the ner_genes_phenotypes and assertion_genomic_abnormality_wip models is transforming how gene and phenotype-related entities are extracted and understood from unstructured biomedical text. The NER model identifies critical entities like genes, mutations, and phenotypic diseases, while the assertion model adds context by classifying them into categories: Normal, Affected, and Variant, resulting in structured datasets that are both detailed and meaningful.
This combination provides researchers and clinicians with deeper insights, enabling them to identify normal genes, those affected by mutations, and specific genetic variants. These insights are essential for understanding disease progression, finding therapeutic targets, and driving advancements in precision medicine. These models play a key role in personalized care by linking genomic data to tailored treatment plans and aiding drug discovery by identifying molecular targets associated with genetic conditions.
Automating the extraction and contextualization of entities simplifies genomic workflows, reduces manual errors, and improves overall efficiency. By converting unstructured data into actionable insights, these models foster innovation in genomics, enhance patient care, and support better decision-making in healthcare and biomedical research.






























