The RegexMatcherInternal class leverages the power of regular expressions to identify and associate specific patterns within text data with predefined entities, such as dates, SSNs, and email addresses. This method facilitates targeted entity extraction by matching text patterns to specified entities.
In the realm of natural language processing, the ability to extract entities from text data plays a crucial role in various applications. In addition to the various Named Entity Recognition (NER) models ( SparkNLP MedicalNerModel using Bidirectional LSTM-CNN architecture and BertForTokenClassification ) in our Models Hub, our library offers powerful rule-based annotators such as ContextualParser, TextMatcher, RegexMatcher, and EntityRuler. In this blog post, we will delve into a powerful annotator known as the RegexMatcherInternal class that assists in this process. This class implements an internal annotator approach to match a set of regular expressions with provided entities, enabling users to associate specific patterns within text data with predetermined entities, such as dates, mentioned within the text.
This post covers the importance of understanding how to match exact phrases by using regex patterns and becoming comfortable with the different parameters of the RegexMatcherInternal. By mastering these skills, users can enhance their text data analysis capabilities and efficiently extract structured information from unstructured or semi-structured textual data. Regex patterns offer a powerful method for identifying specific patterns within text data, while the parameters of the RegexMatcherInternal provide flexibility in controlling the matching strategy. By delving into the intricacies of regex patterns and the RegexMatcherInternal, users can streamline their entity recognition processes and optimize their text data analysis workflows.
Let’s begin with a brief Spark NLP introduction and then delve into the specifics of RegexMatcherInternal usage.
John Snow Labs – Medical Language Models
The Healthcare Library is a powerful component of John Snow Labs’ Spark NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,200 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Spark NLP and related tools.
John Snow Labs also offers periodic certification training to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
Setting Up the John Snow Labs’ Healthcare NLP & LLM library
To set up the John Snow Labs’ Healthcare NLP & LLM library, follow the instructions provided here.
Additionally, you can refer to the Healthcare NLP GitHub repository, which includes sample notebooks. Each notebook contains an initial part that demonstrates how to set up Healthcare NLP on Google Colab, under a section named “Colab Setup”.
RegexMatcherInternal
Advanced Regex Matching for Entity Recognition in Healthcare NLP
The RegexMatcherInternal class in Spark NLP is a robust tool designed to match regular expressions with specific entities in text data. This is particularly useful for tasks like identifying dates, names, or other patterns within unstructured text.
Key Features
- Customizable Rules: Define your own regex rules paired with entities.
- Flexible Input: Set rules directly using the
setRulesmethod or load them from an external file withsetExternalRules. - Matching Strategies: Choose from
MATCH_FIRST,MATCH_ALL, orMATCH_COMPLETEto control how matches are processed. - Annotation Types: Supports
DOCUMENTas input and outputs asCHUNK.
!mkdir -p rules
rules = '''
(\d{1,3}\.){3}\d{1,3}~IPADDR
\d{4}-\d{2}-\d{2}|\d{2}/\d{2}/\d{2}|\d{2}/\d{2}/\d{2}~DATE
'''
with open('./rules/regex_rules.txt', 'w') as f:
f.write(rules)
document_assembler = nlp.DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
regex_matcher_internal = medical.RegexMatcher()\
.setInputCols('document')\
.setOutputCol("regex_matches")\
.setExternalRules(path='./rules/regex_rules.txt', delimiter='~')
nlpPipeline = nlp.Pipeline(
stages=[
document_assembler,
regex_matcher_internal
])
In the provided Pegex patterns above:
(\d{1,3}\.){3}\d{1,3}~IPADDR: This pattern represents an IP address in the format of###.###.###.###, where each###can be 1 to 3 digits. It is tagged as IPADDR.\d{4}-\d{2}-\d{2}|\d{2}/\d{2}/\d{2}|\d{2}/\d{2}/\d{2}~DATE: This pattern matches dates in different formats –YYYY-MM-DD,MM/DD/YY, orMM/DD/YY. It is tagged as DATE.
The code writes these patterns to a file named regex_rules.txt in a folder named rules. We are giving these rules inside the pipeline by using the setExternalRules parameter of RegexMatcherInternal.
Let’s use a sample text to evaluate the performance of the RegexMatcherInternal annotator. After that, we will proceed to fit and transform the dataframe to extract the results.
text = """Name : Hendrickson, Ora, Record date: 2093-01-13, MR #719435.
Dr. John Green, ID: 1231511863, IP 203.120.223.13
He is a 60-year-old male was admitted to the Day Hospital for cystectomy on 01/13/93
Patient's VIN : 1HGBH41JXMN109286, SSN #333-44-6666, Driver's license no: A334455B.
Phone (302) 786-5227, 0295 Keats Street, San Francisco, E-MAIL: smith@gmail.com."""
data = spark.createDataFrame([[text]]).toDF("text")
result = nlpPipeline.fit(data).transform(data)
Export the results to a dataframe with the columns — matched dates and the ner label :
result_df = result.select(F.explode(F.arrays_zip(result.regex_matches.result,
result.regex_matches.begin,
result.regex_matches.end,
result.regex_matches.metadata)).alias("cols"))\
.select(F.expr("cols['0']").alias("regex_result"),
F.expr("cols['1']").alias("begin"),
F.expr("cols['2']").alias("end"),
F.expr("cols['3']['entity']").alias("ner_label"))
result_df.show()
Output:
+--------------+-----+---+---------+ | regex_result|begin|end|ner_label| +--------------+-----+---+---------+ | 2093-01-13| 38| 47| DATE| |203.120.223.13| 97|110| IPADDR| | 01/13/93| 188|195| DATE| +--------------+-----+---+---------+
Crafting Your Own Pipeline: Unleashing the Power of RegexMatcherInternal
As a versatile tool, you have the ability to create your own pipeline tailored to your specific use case. This pipeline can incorporate multiple models, including those created with the powerful RegexMatcherInternal.
In this blog post, we will explore one of the key components within the pipeline — the mail_regex_parser_model.
rule_path = "rules/mail_regex_rule.txt"
model_path = "regex_models/mail_regex_parser_model"
with open(rule_path, 'w') as f:
f.write("""[A-Za-z0-9._%+-]+@[A-Za-z0-9.-]+\.[A-Za-z]{2,}~EMAIL""")
mail_regex_matcher = medical.RegexMatcher()\
.setExternalRules(rule_path, "~") \
.setInputCols(["document"]) \
.setOutputCol("mail_matched_text") \
.setStrategy("MATCH_ALL")
regex_parser_pipeline = nlp.Pipeline(
stages=[
document_assembler,
mail_regex_matcher
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
regex_parser_model = regex_parser_pipeline.fit(empty_data)
regex_parser_model.stages[-1].write().overwrite().save(model_path)
By understanding the functionality and implementation of the mail_regex_parser_model, you’ll be equipped with the knowledge to build robust and customized pipelines that cater to your unique requirements.
document_assembler = nlp.DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentence_detector = nlp.SentenceDetectorDLModel.pretrained("sentence_detector_dl_healthcare","en","clinical/models")\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = nlp.Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
regex_matcher_ssn = medical.RegexMatcherModel.load("regex_models/ssn_regex_parser_model")\
.setInputCols(["sentence"]) \
.setOutputCol("ssn_matched_text")
regex_matcher_age = medical.RegexMatcherModel.load("regex_models/age_regex_parser_model")\
.setInputCols(["sentence"]) \
.setOutputCol("age_matched_text")
regex_matcher_mail = medical.RegexMatcherModel.load("regex_models/mail_regex_parser_model")\
.setInputCols(["sentence"]) \
.setOutputCol("mail_matched_text")
regex_matcher_phone = medical.RegexMatcherModel.load("regex_models/phone_regex_parser_model")\
.setInputCols(["sentence"]) \
.setOutputCol("phone_matched_text")
chunk_merge = medical.ChunkMergeApproach()\
.setInputCols("ssn_matched_text",
"age_matched_text",
"mail_matched_text",
"phone_matched_text")\
.setOutputCol("ner_chunk")\
.setMergeOverlapping(True)\
.setChunkPrecedence("field")
nlpPipeline = nlp.Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
regex_matcher_ssn,
regex_matcher_age,
regex_matcher_mail,
regex_matcher_phone,
chunk_merge
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
regex_pipeline_model = nlpPipeline.fit(empty_data)
light_model = nlp.LightPipeline(regex_pipeline_model)
text = """Name : Hendrickson, Ora, Record date: 2093-01-13, MR #719435.
Dr. John Green, ID: 1231511863, IP 203.120.223.13.
He is a 60-year-old male was admitted to the Day Hospital for cystectomy on 01/13/93.
Patient's VIN : 1HGBH41JXMN109286, SSN #333-44-6666, Driver's license no: A334455B.
Phone (302) 786-5227, 0295 Keats Street, San Francisco, E-MAIL: smith@gmail.com."""
result = light_model.fullAnnotate(text)
ner_chunk = []
ner_label = []
begin = []
end = []
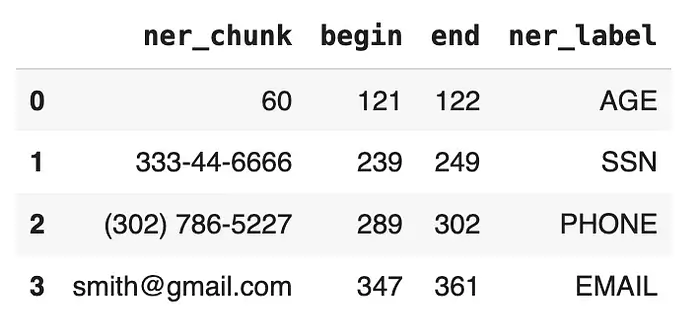
for n in result[0]['ner_chunk']:
begin.append(n.begin)
end.append(n.end)
ner_chunk.append(n.result)
ner_label.append(n.metadata['entity'])
import pandas as pd
df = pd.DataFrame({'ner_chunk':ner_chunk, 'begin': begin, 'end':end,
'ner_label':ner_label})
df
Output:
You can save a trained Spark NLP pipeline model to disk. This allows you to persist the model and reuse it in the future, without having to go through the training process again.
regex_pipeline_model.write().overwrite().save("regex_pipeline_model")
Loading the previously saved pipeline model from disk.
from sparknlp.pretrained import PretrainedPipeline
regex_pipeline_loaded = PretrainedPipeline.from_disk("regex_pipeline_model")
For extended examples of usage, see the Spark NLP Workshop Repository.
Reference Documentation: RegexMatcherInternal
Conclusion
Information extraction through regular expressions is a fundamental aspect of Natural Language Processing (NLP) that plays a vital role in transforming raw text into structured data. The RegexMatcherInternal tool in Healthcare NLP allows users to create custom regular expressions for extracting specific patterns from text, offering a high level of flexibility for diverse use cases. This capability not only streamlines the extraction of valuable insights but also enhances automation and decision-making processes in healthcare by enabling precise identification of relevant information.
By integrating powerful regular expression capabilities into a scalable NLP framework like Spark NLP, the extraction of insights from text in healthcare becomes more efficient and effective, even with large datasets. The RegexMatcherInternal in Spark NLP provides a rule-based approach that enables users to define and apply custom regex rules, empowering them to accurately extract specific patterns from text data. This solution is particularly beneficial in healthcare settings where precision and accuracy are crucial for data analysis, decision-making, and improving overall patient care outcomes, making it a versatile and invaluable tool for driving innovation and efficiency in the healthcare domain.































