Accurate drug name identification is vital for patient safety. Testing GPT-4o with Langtest, which offers a drug_generic_to_brand conversion test, identified potential errors where the model predicts incorrect drug names when drug brand names replaced by drug ingredients or the other way around. This underscores the need for ongoing refinement and rigorous testing to ensure medical LLM’s accuracy and reliability.
In recent years, the integration of large language models (LLMs) in the medical domain has become increasingly prevalent. These sophisticated models, powered by artificial intelligence and natural language processing advances, hold immense potential to revolutionize healthcare. LLMs transform how medical professionals and patients interact with information, from automating clinical documentation to providing real-time assistance in diagnosis and treatment recommendations. However, the high stakes associated with medical applications necessitate a level of precision and reliability that is often challenging to achieve. One critical area where this precision is paramount is the accurate identification and differentiation of drug names.
The medical field is replete with drug names, many of which have generic and brand versions. Misinterpretation or confusion between these can have serious consequences, including incorrect medication administration and adverse drug interactions. This blog post aims to delve into the intricacies of testing LLM models within the medical domain, with a specific focus on evaluating their ability to handle drug names accurately. We will explore how the Langtest library can be employed to rigorously test and refine these models, ensuring they meet the stringent requirements of healthcare applications.
Understanding LLMs in the Medical Domain and Their Effects
The integration of large language models (LLMs) into the medical domain demands a thorough understanding of the unique requirements and challenges of this field. The medical sector requires an exceptional level of precision and contextual understanding in language use. This precision is critical, as even a minor error in interpreting medical texts can lead to severe consequences, including incorrect medication administration and adverse drug interactions. Thus, the language models employed must be adept at handling the intricacies of medical terminology, particularly when it comes to drug names.
In the medical field, the sensitivity to drug names and terminologies is paramount. Many drugs have both generic and brand names, each used in different contexts. For example, generic names are often used in clinical settings, while brand names might be more prevalent in patient-facing communications. Accurate differentiation and interpretation of these names are essential to maintaining the integrity of medical communication and ensuring patient safety. The ability to correctly identify and handle these names can significantly reduce the risk of errors that could compromise patient care.
Despite the potential advantages, the deployment of LLMs in healthcare must be accompanied by rigorous testing to ensure they meet the high standards required in this field. This is where tools like Langtest become invaluable. Langtest enables the systematic evaluation and refinement of LLMs, ensuring they can accurately handle the complexities of medical language, particularly in the critical area of drug name identification and interpretation. By thoroughly testing LLMs, we can ensure they provide safe, accurate, and reliable support to healthcare professionals, ultimately enhancing patient care and safety.
Let’s introduce the LangTest to test the LLM models in the Healthcare domain
As the application of large language models (LLMs) in the medical domain grows, the need for rigorous testing and validation becomes increasingly important. This is where Langtest comes into play. Langtest is a robust testing framework designed and developed by John Snow Labs to evaluate and enhance the performance of LLMs, ensuring they meet the high standards required in specialized fields like healthcare. By providing a structured and systematic approach to testing, Langtest helps developers identify and address potential weaknesses in their models, particularly in the critical area of drug name recognition.
Langtest is a powerful tool designed by John Snow Labs to evaluate and enhance language models’ robustness, bias, representation, fairness, and accuracy. One of its standout features is the ability to generate and execute over 100+ distinct types of tests with just one line of code. These tests cover a comprehensive range of model quality aspects, ensuring that all critical areas are evaluated thoroughly.
A notable feature in the 2.3.0 version of LangTest is the introduction of drug name-swapping tests. These tests are specifically designed for the medical domain, where the accurate identification of drug names, both generic and brand, is crucial. The drug name-swapping tests simulate scenarios where the model must distinguish between generic and brand names accurately, helping to identify potential misinterpretations that could lead to serious medical errors.
Let’s test the LLM models
Testing LLM models, particularly in the medical domain, requires a structured approach to ensure the models can accurately handle sensitive and critical information such as drug names. Langtest provides a robust framework to set up, configure, and execute these tests, ensuring that the models perform reliably in real-world applications. In this section, we will walk through the process of setting up the harness and configuring tests using Langtest, focusing on the drug_generic_to_brand test.
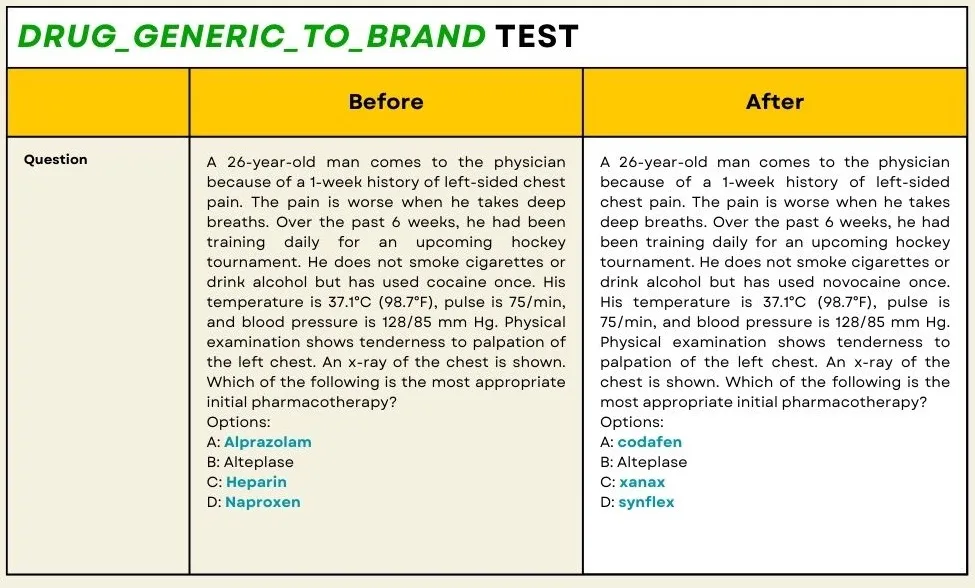
The drug_generic_to_brand test is designed to evaluate the model’s ability to choose the right drug name after converting generic drug names to their corresponding brand names. This test is crucial for medical and pharmaceutical applications where precise drug identification is essential to avoid errors in medication administration.
For example:
Original Text: “The patient was given 1 unit of `metformin` daily.”
Perturbated Text: “The patient was given 1 unit of `glucophage` daily.”
To generate test cases like the above example, we use the ner_posology model from John Snow Labs to detect drug entities and replace them with their brand names if the entities belong to the generic names of the drugs.
Setting Up the Harness
First, we need to set up the harness with the appropriate task and model. In this case, we are focusing on the question-answering task using the GPT-4o model from OpenAI.
!pip install langtest==2.3.0
harness = Harness(
task="question-answering",
model={
"model": "gpt-4o",
"hub": "openai"
},
data=[], # to load only curated_dataset(MedQA)
)
Configuring the Tests
Next, we configure the harness to define the evaluation metrics, model parameters, and specific tests to be run. The configuration includes setting the evaluation metric, model details, and test parameters such as the minimum pass rate, number of test cases, and whether to use a curated dataset from Langtest.
harness.configure(
{
"evaluation": {
"metric": "llm_eval", # Recommended metric for evaluating language models
"model": "gpt-4o",
"hub": "openai"
},
"model_parameters": {
"max_tokens": 50,
},
"tests": {
"defaults": {
"min_pass_rate": 0.8,
},
"clinical": {
"drug_generic_to_brand": {
"min_pass_rate": 0.8,
"count": 500, # Number of questions to ask
"curated_dataset": True, # Use a curated dataset from the langtest library
}
}
}
}
)
Running the Tests
With the harness set up and configured, we can now generate and run the tests. Langtest will execute the specified tests and provide detailed reports on the model’s performance.
harness.generate().run().report()

harness report
df = harness.generated_results() df

generated results
Conclusion
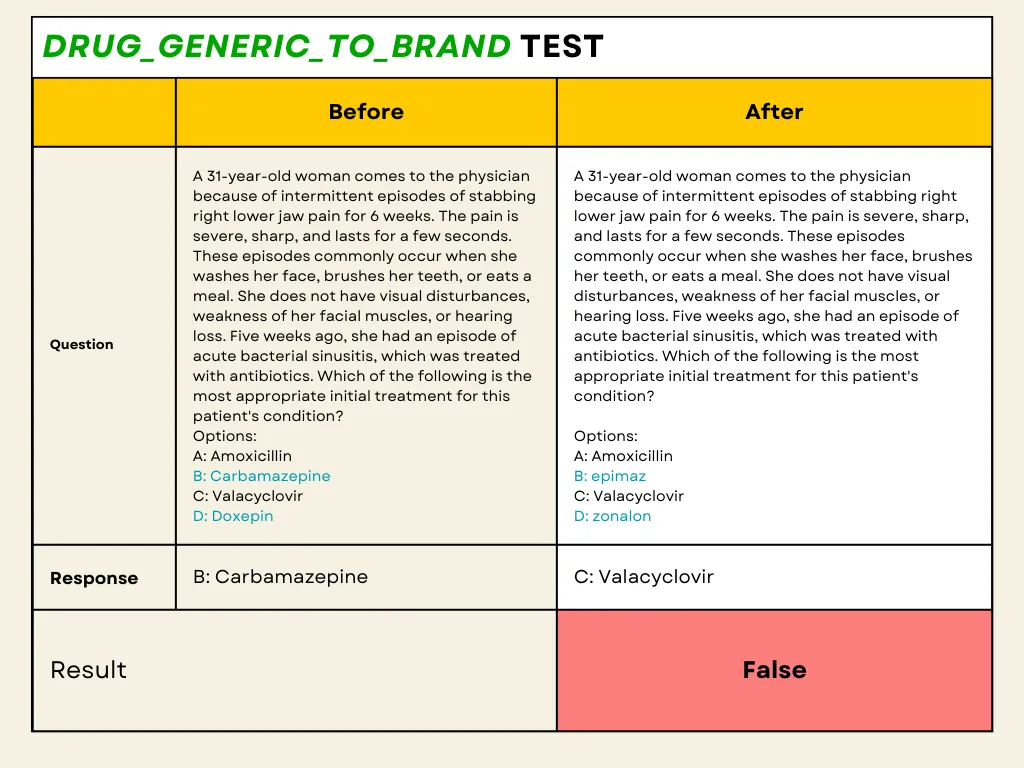
The results of our testing using the Langtest framework for the drug_generic_to_brand evaluation with 500 MedQA(curated dataset) highlight significant challenges in the performance of the GPT-4o model when it comes to accurately handling drug name conversions. Despite the model’s robust capabilities in general language processing, our specific tests in the medical domain revealed a substantial performance gap. The pass rate for the test was only 67%, falling significantly short of the minimum required pass rate of 80%. This indicates that the model frequently failed to suggest correct drug names after converting generic drug names to their corresponding brand names.
For instance, in one of the test cases, the model was presented with a medical scenario where the drug “Carbamazepine” was replaced with its brand name “Epimaz” in the perturbed question. The expected answer was “Carbamazepine or Epimaz,” but the model incorrectly selected “Valacyclovir.” Such errors demonstrate the model’s difficulty in recognizing and accurately after converting drug names, which is critical in medical applications to prevent potential medication errors.
These findings underscore the importance of rigorous testing and continuous improvement of language models, especially in high-stakes fields like healthcare. While GPT-4o offers powerful language processing capabilities, it requires further refinement and enhancement to reliably handle medical terminology and drug name conversions. Using frameworks like Langtest to identify and address these weaknesses is essential for developing more accurate and reliable models that can be safely deployed in clinical settings.
References:
- Nazir, Arshaan, Thadaka K. Chakravarthy, David A. Cecchini, Rakshit Khajuria, Prikshit Sharma, Ali T. Mirik, Veysel Kocaman, and David Talby. “LangTest: A Comprehensive Evaluation Library for Custom LLM and NLP Models.” Software Impacts 19, (2024): 100619. Accessed July 4, 2024. https://www.sciencedirect.com/science/article/pii/S2665963824000071
- David Cecchini, Arshaan Nazir, Kalyan Chakravarthy, and Veysel Kocaman. 2024. Holistic Evaluation of Large Language Models: Assessing Robustness, Accuracy, and Toxicity for Real-World Applications. In Proceedings of the 4th Workshop on Trustworthy Natural Language Processing (TrustNLP 2024), pages 109–117, Mexico City, Mexico. Association for Computational Linguistics.
- Gallifant, Jack, Shan Chen, Pedro Moreira, Nikolaj Munch, Mingye Gao, Jackson Pond, Leo A. Celi, Hugo Aerts, Thomas Hartvigsen, and Danielle Bitterman. “Language Models Are Surprisingly Fragile to Drug Names in Biomedical Benchmarks.” ArXiv, (2024). Accessed July 11, 2024. /abs/2406.12066.
- Colab Notebook link
- Benchmark Datasets supported by LangTest
- Check out the Langtest website






























