Introduction
In the world of healthcare and medical research, the ability to access and share medical images is crucial for diagnosis, treatment, and scientific investigation. However, these images often contain sensitive patient information, making their handling delicate. De-identification of DICOM files is a critical process that ensures patient privacy while enabling the secure exchange of medical images. This post will explore how Visual NLP can be a game-changer for de-identifying DICOM files at scale.
DICOM stands for “Digital Imaging and Communications in Medicine.” It is a standard for transmitting, storing, and sharing medical images and associated information in the field of healthcare. DICOM is widely used in medical imaging technologies such as X-rays, MRIs, CT scans, and ultrasounds to ensure interoperability and consistency in handling medical data across different devices and systems.
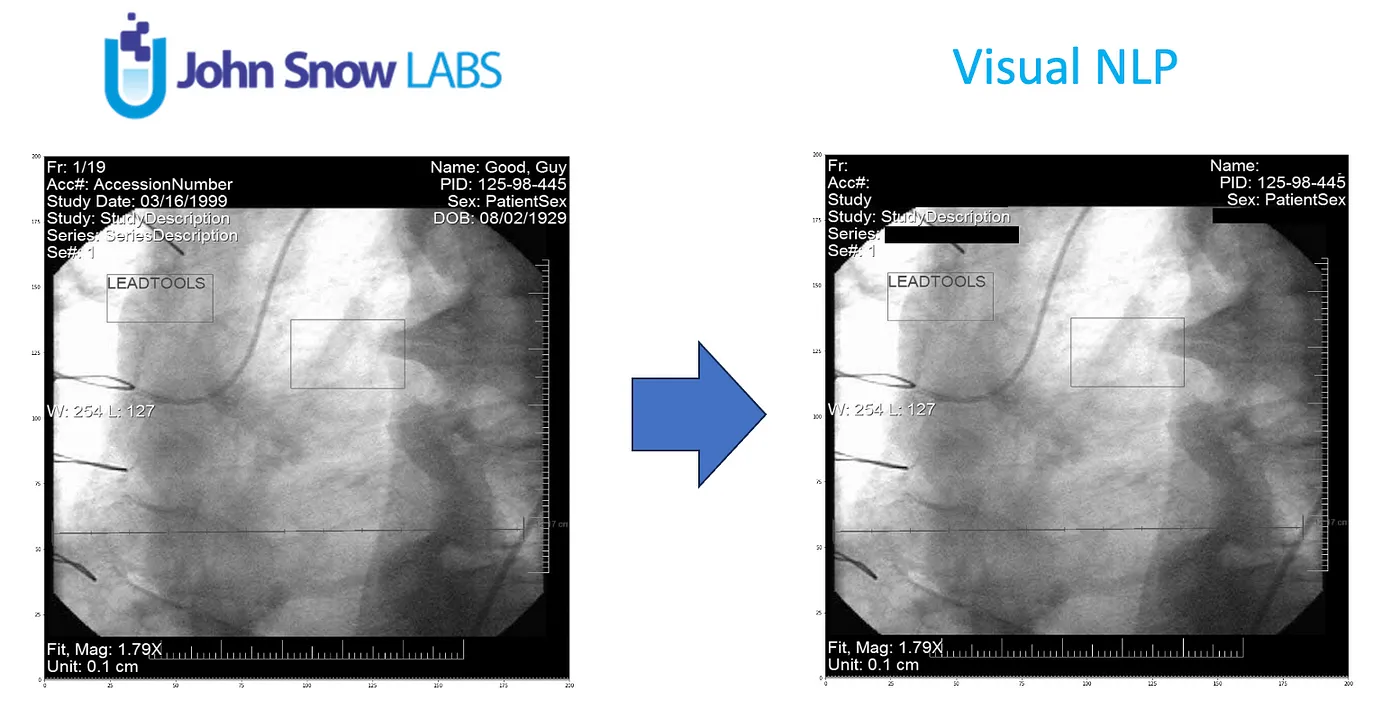
However, these images are not just pictures; they contain a wealth of metadata containing sensitive information, such as patient names, birthdates, and medical record numbers (PHI).
PHI is sensitive and confidential information, and its presence in DICOM requires careful handling to ensure patient privacy and compliance with healthcare privacy regulations, such as HIPAA (Health Insurance Portability and Accountability Act) in the United States.
De-identification is the process of removing or anonymizing this sensitive information while preserving the diagnostic value of the medical images.
The Challenge of De-Identification at Scale
As medical institutions and research facilities accumulate vast amounts of medical imaging data, the need for efficient and scalable de-identification solutions becomes evident. Manually de-identifying each image is time-consuming, error-prone, and often impractical when dealing with large datasets.
This is where Visual NLP steps in.
The Power of Visual NLP
Visual NLP is an advanced tool built on the Apache Spark framework, designed to handle optical character recognition (OCR) tasks, including DICOM de-identification, at scale. Here’s how it can revolutionize the process:
Scalability: Visual NLP leverages the distributed computing power of Apache Spark, making it capable of processing vast quantities of DICOM files in parallel. This scalability is essential when dealing with big data in healthcare.
Automation: It automates the de-identification process, reducing the need for manual intervention. This speeds up the workflow and minimizes the risk of human error.
Customization: Visual NLP allows for the creation of custom de-identification rules to meet specific institutional or regulatory requirements. You can define what information to redact or anonymize based on your needs.
Accuracy: The OCR capabilities of Visual NLP ensure accurate text extraction from DICOM images, enabling precise identification and removal of sensitive data.
De-Identification Dicom
PHI data can be present in Dicom files in several places:
- metadata
- pixel data as burned in pixels
- overlay data as annotations
- encapsulated documents
Let’s discuss more detail each item.
Metadata
DICOM metadata contains a lot of patient-related information.
Here are some common examples of PHI that can be found in DICOM metadata:
- Patient Name: The patient’s full name or other identifying information.
- Patient ID: A unique identifier for the patient, including a medical record number or other patient-specific code.
- Date of Birth: The patient’s date of birth can be used to confirm the patient’s identity.
- Study Date: The date on which the medical study (e.g., an MRI or CT scan) was performed.
- Accession Number: A unique identifier assigned to a specific study or examination.
- Institution Name: The name of the healthcare institution where the study was conducted.
- Referring Physician: The name of the physician who referred the patient for the study.
- Performing Physician: The name of the physician who performed the study.
- Patient History: Any medical history or clinical information related to the patient’s condition.
- Patient’s Sex: The gender of the patient.
Pixel data
“burned-in pixel data” refers to image information that has been permanently embedded or “burned” into the image itself, making it an integral part of the pixel data. This means the information cannot be easily removed or modified because it is part of the image’s raw data.
Overlay data
“overlay data” refers to additional graphical or textual information that can be superimposed or overlaid onto medical images. This overlay data provides annotations, measurements, or other information types that enhance the image’s interpretation and understanding.
Overlay data is organized into overlay planes. Each overlay plane represents a separate layer of graphical or textual information that can be superimposed on the main image. Multiple overlay planes can be used to add different types of annotations or information to an image.
Encapsulated Documents
Encapsulated Documents in DICOM are used to associate textual or document information with medical images. For example, they can include clinical reports, patient histories, annotations, or other textual data relevant to the medical images.
DICOM Encapsulated Documents can contain various document formats, such as PDF (Portable Document Format), HTML (Hypertext Markup Language), or plain text. The format used is typically specified within the DICOM object.
Example of the Dicom document with PHI in metadata and in pixel data
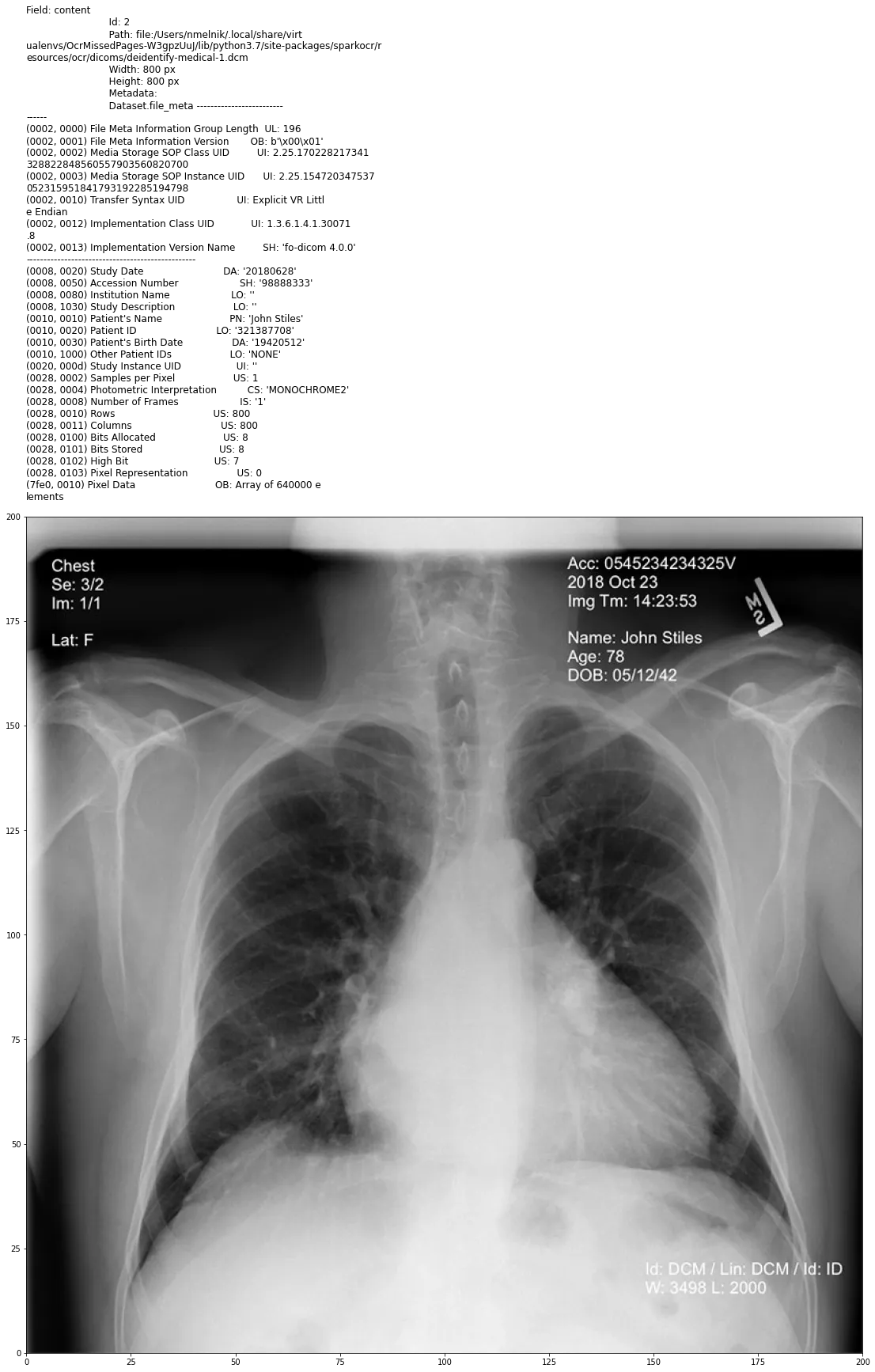
In the next post, we start to deal with DICOM files using Visual NLP capabilities.