In this post, we will explain how to use the new Spark NLP‘s pre-trained NER NLP model ner_radiology that can identify entities related to radiology.
Introduction
Named Entity Recognition (NER) is a dynamic field of Natural Language Processing (NLP) research and is defined as an automatic system to detect “entities” in a text. These entities depend on the application scenario and can be People, Location, and Organization for a generic news article text, or, as in the case here, entities related to radiology.
The capability to automatically identify radiology entities can help health practitioners to structure and organize big data sets at scale. Usually, without an automatic model, practitioners need to manually label the entities in the text if they want to analyze the data they have or create a model that uses this information as input.
John Snow Labs’ Spark NLP
Spark NLP is an open-source Java and Python NLP library built on top of Apache Spark from John Snow Labs. It is one of the top growing NLP production-ready solutions with support for all main tasks related to NLP in more than 46 languages.
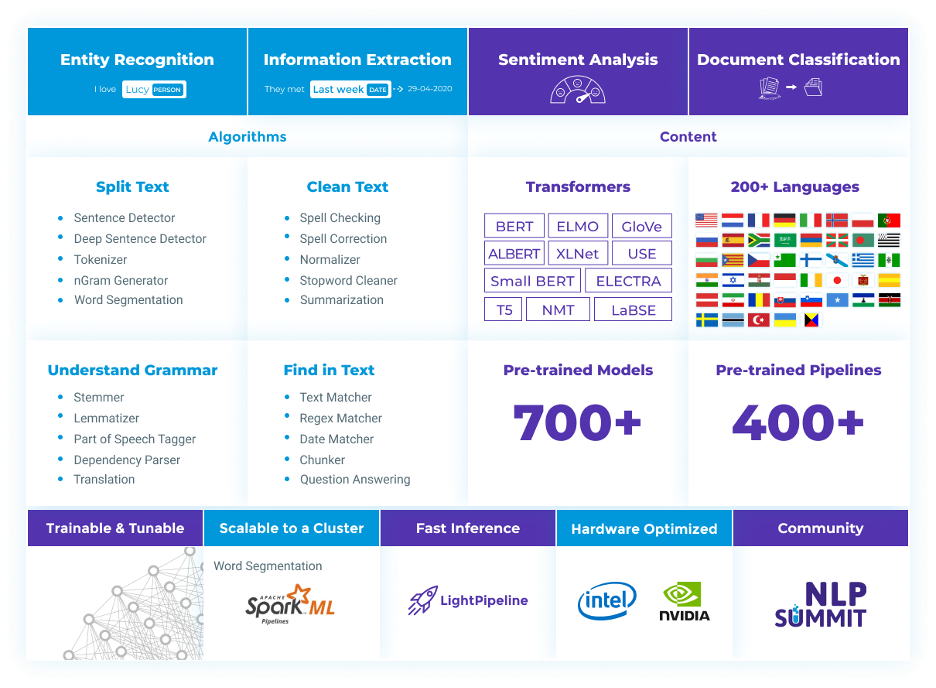
Figure 1: Open-Source Spark NLP
Apart from the open-source library, the company maintains the licensed library specialized for healthcare solutions, SPARK NLP for Healthcare.
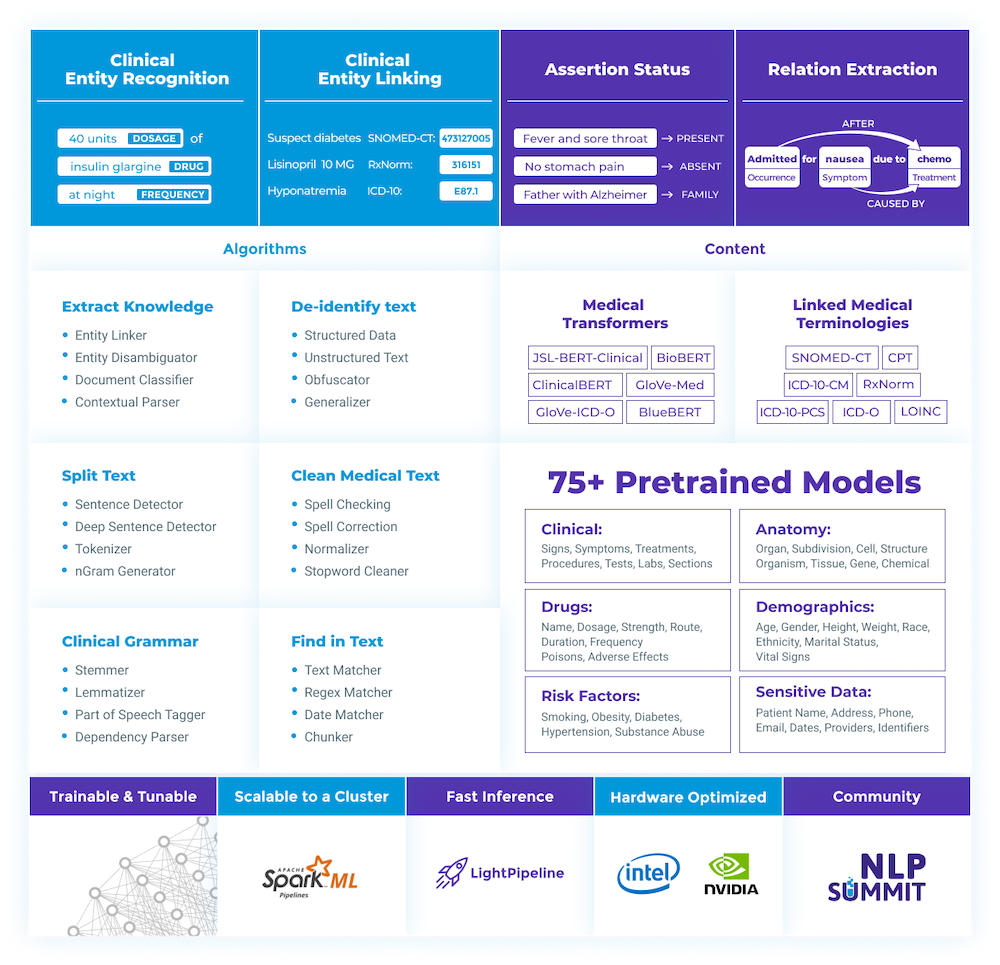
Spark NLP for Healthcare contains a pre-trained model for Named Entity Recognition in the scope of radiology.
Spark NLP pipeline
Spark NLP is built on top of Apache Spark and is based on the concept of pipelines, where we can perform several NLP tasks in our data with a unified pipeline. For an introduction of Spark NLP functionalities, refer to [1].
Here, we will explore the Spark NLP for Healthcare capabilities for Radiology NER using the new model.
The new ner_radiology model was trained on a custom dataset comprising of MIMIC-CXR [5] and MT Radiology texts, and can identify the following entities:
- ImagingTest: Name of an imaging test (Ultrasound)
- Imaging_Technique: Name of the technique used
- ImagingFindings: Diagnostic or identifications found on tests (Ovoid mass)
- OtherFindings: Other findings
- BodyPart: Name of anatomic part of the body (Shoulder)
- Direction: Detail direction of the image test (Bilateral
- Test: Name of the test (not imaging)
- Symptom: Symptom identified
- Disease_Syndrome_Disorder: Name of health condition (Lipoma)
- Medical_Device: Name of medical devices
- Procedure: Name of procedure
- Measurements: Measurements such as 0.5 x 0.4
- Units: Units such as cm, ml
In Spark NLP, we build a pipeline containing all the steps (stage) of the model, and in the case of NER models, we need to transform the text into a document annotator (DocumentAssembler), identify sentences (SentenceDetector), tokenize (Tokenizer), transform to embeddings vectors (WordEmbeddingsModel), identify entities (NerModel) and finally convert the entities to standard formats (NerConverter). The full pipeline in Python can be seen below.
# Import modules from pyspark.ml.pipeline import PipelineModel from sparknlp.base import DocumentAssembler from sparknlp.annotator import SentenceDetector, Tokenizer, WordEmbeddingsModel from sparknlp.annotator import NerDLModel, NerConverter document_assembler = DocumentAssembler() \ .setInputCol("text") \ .setOutputCol("document") sentence_detector = SentenceDetector()\ .setInputCols(["document"])\ .setOutputCol("sentence") tokenizer = Tokenizer()\ .setInputCols(["sentence"])\ .setOutputCol("token") embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical","en","clinical/models")\ .setInputCols("document", "token") \ .setOutputCol("embeddings") ner = NerDLModel.pretrained("ner_radiology", "en", "clinical/models") \ .setInputCols(["document", "token", "embeddings"]) \ .setOutputCol("ner") ner_converter = NerConverter() \ .setInputCols(["document", "token", "ner"]) \ .setOutputCol("ner_chunk") pipeline = Pipeline(stages=[ document_assembler, sentence_detector, tokenizer, embeddings, ner, ner_converter])
We then can initialize the pipeline on an example:
Bilateral breast ultrasound was subsequently performed, which demonstrated an ovoid mass measuring approximately 0.5 x 0.5 x 0.4 cm in diameter located within the anteromedial aspect of the left shoulder. This mass demonstrates isoechoic echotexture to the adjacent muscle, with no evidence of internal color flow. This may represent benign fibrous tissue or a lipoma.
To do that, we will use two pipeline objects: the Pipeline and the LightPipeline. The first one is useful to keep all the stages in one single object and is used on spark data frames, while the second one is useful for fast predictions and can be used directly on strings (or list of strings).
# Send example to spark data frame
example = spark.createDataFrame([['''Bilateral breast ultrasound was subsequently performed, which
demonstrated an ovoid mass measuring approximately 0.5 x 0.5 x 0.4 cm in diameter located within the
anteromedial aspect of the left shoulder. This mass demonstrates isoechoic echotexture to the
adjacent muscle, with no evidence of internal color flow. This may represent benign fibrous tissue
or a lipoma.''']]).toDF("text")
# Initialize the model
model = nlpPipeline.fit(example)
# Make predictions on the example (full model)
result = model.transform(example)
# Create a light model for fast predictions
lmodel = LightPipeline(model)
# Use texts directly
result_light = lmodel.fullAnnotate("Bilateral breast ultrasound was subsequently performed, which
demonstrated an ovoid mass measuring approximately 0.5 x 0.5 x 0.4 cm in diameter located within
the anteromedial aspect of the left shoulder. This mass demonstrates isoechoic echotexture to
the adjacent muscle, with no evidence of internal color flow. This may represent benign fibrous
tissue or a lipoma."
To visualize the results of the light model, we can run:
for res in result_light[0]['ner_chunk']:
print(f"Chunk: {res.result}, Entity: {res.metadata['entity']}")
Resulting:
Chunk: Bilateral breast, Entity: BodyPart Chunk: ultrasound, Entity: ImagingTest Chunk: ovoid mass, Entity: ImagingFindings Chunk: 0.5 x 0.5 x 0.4, Entity: Measurements Chunk: cm, Entity: Units Chunk: anteromedial aspect of the left shoulder, Entity: BodyPart Chunk: mass, Entity: ImagingFindings Chunk: isoechoic echotexture, Entity: ImagingFindings Chunk: muscle, Entity: BodyPart Chunk: internal color flow, Entity: ImagingFindings Chunk: benign fibrous tissue, Entity: ImagingFindings Chunk: lipoma, Entity: Disease_Syndrome_Disorder
And for the full model:
result.select(F.explode(F.arrays_zip('ner_chunk.result', 'ner_chunk.metadata')).alias("cols")) \
.select(F.expr("cols['0']").alias("chunk"),
F.expr("cols['1']['entity']").alias("entities"))\
.show(truncate=False)
Resulting:
| chunk | entities |
|---|---|
| Bilateral | Direction |
| breast | BodyPart |
| ultrasound | ImagingTest |
| ovoid mass | ImagingFindings |
| 0.5 x 0.5 x 0.4 | Measurements |
| cm | Units |
| anteromedial aspect | Direction |
| left | Direction |
| shoulder | BodyPart |
| mass | ImagingFindings |
| isoechoic echotexture | ImagingFindings |
| muscle | BodyPart |
| internal color flow | ImagingFindings |
| benign fibrous tissue | ImagingFindings |
| lipoma | Disease_Syndrome_Disorder |
Overall, the model achieves good performance on the entities (except for the OtherFindings that is less relevant), as can be seen in the table below.
| entity | tp | fp | fn | total | precision | recall | f1 |
|---|---|---|---|---|---|---|---|
| OtherFindings | 8 | 15 | 63 | 71 | 0.3478 | 0.1127 | 0.1702 |
| Measurements | 481 | 30 | 15 | 496 | 0.9413 | 0.9698 | 0.9553 |
| Direction | 650 | 137 | 94 | 744 | 0.8259 | 0.8737 | 0.8491 |
| ImagingFindings | 1,345 | 355 | 324 | 1,669 | 0.7912 | 0.8059 | 0.7985 |
| BodyPart | 1,942 | 335 | 290 | 2,232 | 0.8529 | 0.8701 | 0.8614 |
| Medical_Device | 236 | 75 | 64 | 300 | 0.7588 | 0.7867 | 0.7725 |
| Test | 222 | 41 | 48 | 270 | 0.8441 | 0.8222 | 0.8330 |
| Procedure | 269 | 117 | 116 | 385 | 0.6969 | 0.6987 | 0.6978 |
| ImagingTest | 263 | 50 | 43 | 306 | 0.8403 | 0.8595 | 0.8498 |
| Symptom | 498 | 101 | 132 | 630 | 0.8314 | 0.7905 | 0.8104 |
| Disease_Syndrome_Disorder | 1,180 | 258 | 200 | 1,380 | 0.8206 | 0.8551 | 0.8375 |
| Units | 269 | 10 | 2 | 271 | 0.9642 | 0.9926 | 0.9782 |
| Imaging_Technique | 140 | 38 | 25 | 165 | 0.7865 | 0.8485 | 0.8163 |
The model achieves and micro accuracy of 0.8315 and a macro accuracy of 0.7524.
Conclusion
In this post, we introduced a new clinical Named Entity Resolution model that identifies entities related to radiology. If you want to try them on your data, you can ask for a Spark NLP Healthcare free trial license.
Being used in enterprise projects and built natively on Apache Spark and TensorFlow as well as offering all-in-one state-of-the-art NLP solutions, Spark NLP library provides simple, performant as well as accurate NLP notations for machine learning pipelines that can scale easily in a distributed environment.
If you want to find out more and start practicing Spark NLP, please check out the reference resources below.
Further reading
- Introduction to spark NLP
- Text Classification in Spark NLP
- NER with BERT in Spark NLP
- John Snow Labs training materials
- IMIC-CXR: A large publicly available database of labeled chest radiographs. Johnson AEW, Pollard TJ, Berkowitz S, Greenbaum NR, Lungren MP, Deng C-Y, Mark RG, Horng S. arXiv. Available from: https://arxiv.org





























