De-identification is the process of removing or masking sensitive, personally identifiable information from data, particularly in medical records. This is crucial to protect patient privacy and comply with regulations like the General Data Protection Regulation (GDPR) in Europe. While healthcare data is invaluable for research, improper handling can lead to severe privacy risks. Once de-identified, medical data becomes a powerful resource for various purposes such as drug development, disease treatment research, and epidemiological studies. This allows researchers to analyze large datasets without compromising patient confidentiality.
Protected Health Information (PHI) refers to any data that can identify an individual and is tied to their health records. This includes identifiers like DATE, NAME, LOCATION, PROFESSION, AGE, ID, CONTACT when connected to health data. Ensuring that these details are either removed or altered is a key component of de-identification.
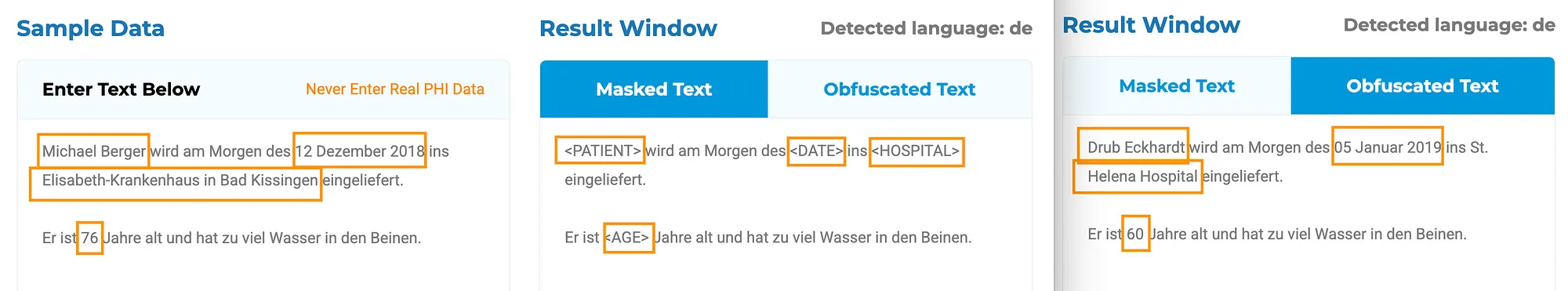
De-identifying medical texts in German poses unique challenges:
- Complex syntactic structures: German has long compound words that can contain identifiable information.
- Domain-specific terminology: Medical jargon varies significantly by language, requiring highly specialized NLP models.
- High density of sensitive data: Medical documents often consolidate critical information in single sections, demanding sophisticated extraction techniques.
Unlike English, which benefits from a broad range of natural language processing (NLP) tools and datasets, German texts are underserved in this area. This makes tailored solutions necessary to overcome these linguistic complexities.
In this blog post, we demonstrated how PHI (Protected Health Information) in German medical texts can be efficiently extracted and de-identified using the Healthcare NLP Library. We discussed the unique challenges posed by German texts, such as complex syntactic structures, domain-specific terminology, and dense sensitive data. Additionally, we showed how to build a custom German de-identification pipeline using pre-trained models, processing stages, and rule-based matchers to enhance accuracy. With flexible deployment options, the library offers a reliable solution for ensuring compliance with GDPR while maintaining data utility for research and analysis.
Introduction to the Healthcare NLP Library
The Healthcare NLP Library is designed to address the challenges of processing medical texts, particularly in non-English languages like German. With over 2,500 specialized models, it helps ensure the secure de-identification of sensitive information in compliance with GDPR, while offering scalability, flexibility, and a range of specialized tools tailored to healthcare needs.
Our solutions are tailored to overcome the complexities of German medical texts, with 51 models optimized specifically for healthcare. Only some of them are using for de-identification purpose. These solutions ensure that clinical data can be de-identified without compromising its utility for research and analysis.
- The library breaks large medical texts into logical sections for easier handling, ensuring complex structures like German compound words are correctly processed.
- It recognizes the context surrounding sensitive terms, helping prevent errors during anonymization.
- The Healthcare NLP Library is designed to handle large-scale data with ease. By employing scalable infrastructure, it can efficiently process millions of records without performance bottlenecks. This scalability is crucial for hospitals and research centers managing vast clinical databases. Additionally, users can deploy and run our models:
- Locally, within a secure environment, which is ideal for maintaining privacy and working in isolated networks without internet access.
- In cloud or enterprise settings, enabling collaboration across research teams while ensuring data security through encrypted environments.
- Ensure GDPR compliance with automated de-identification processes.
- Utilize de-identified data across studies, such as drug development and epidemiology, without the risk of privacy breaches.
- Maintain productivity by significantly reducing manual anonymization tasks, freeing up time for deeper research.
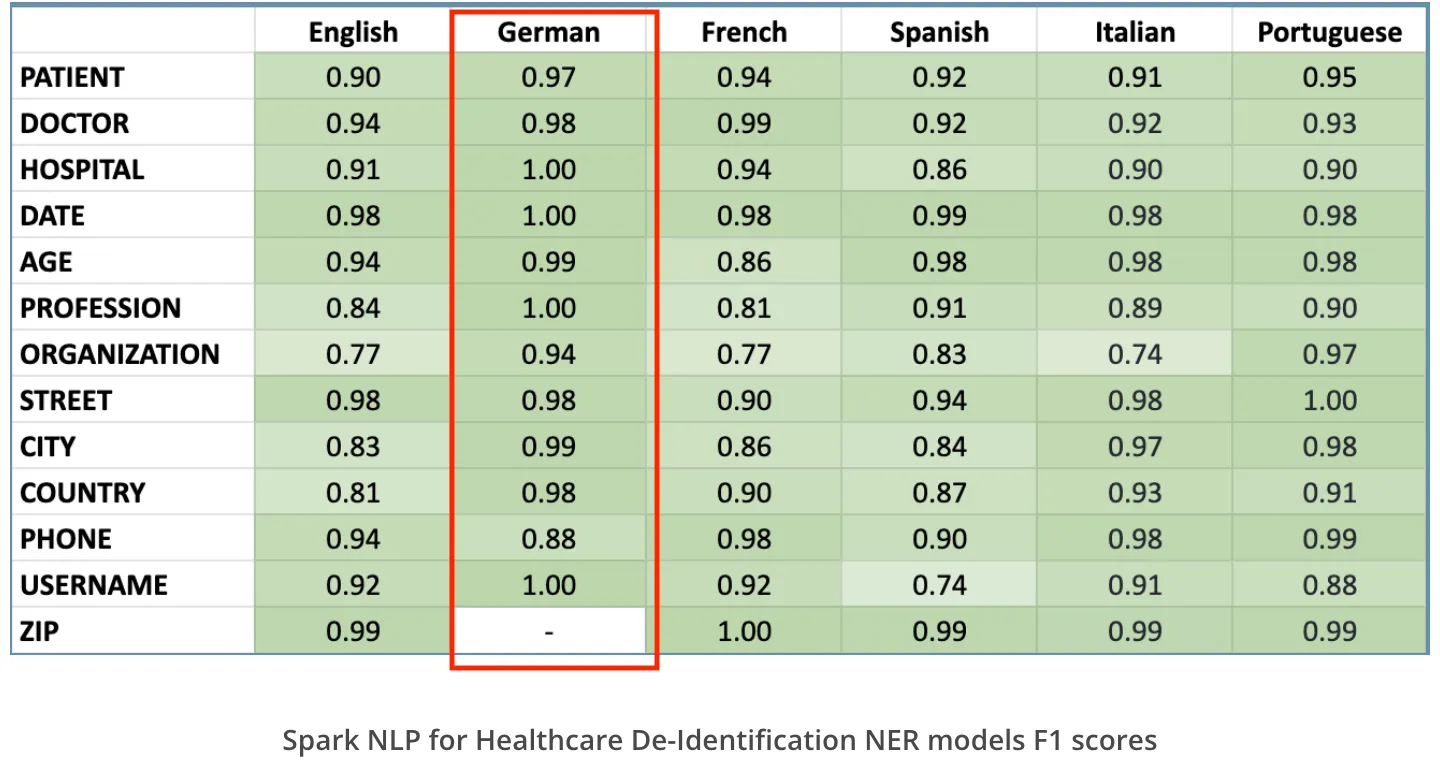
Healthcare NLP has data de-identification tools to de-identify clinical notes in 6 spoken languages. Here are the supported languages and the accuracy metrics for each one of them.
For a more in-depth understanding of de-identification, you can refer to this article.
Explore our interactive notebook and demo for detailed information and visualizations of de-identification workflows. For a deep dive into all available parameters and model capabilities, visit the provided resources.
De-Identification Techniques and Strategies
When dealing with sensitive medical data, de-identification ensures that Protected Health Information (PHI) remains secure. This process involves techniques such as replacing entities with realistic but synthetic ones while maintaining the consistency of entity formats. By preserving structural integrity, de-identified data remains useful for research, AI training, and analytics without compromising privacy.
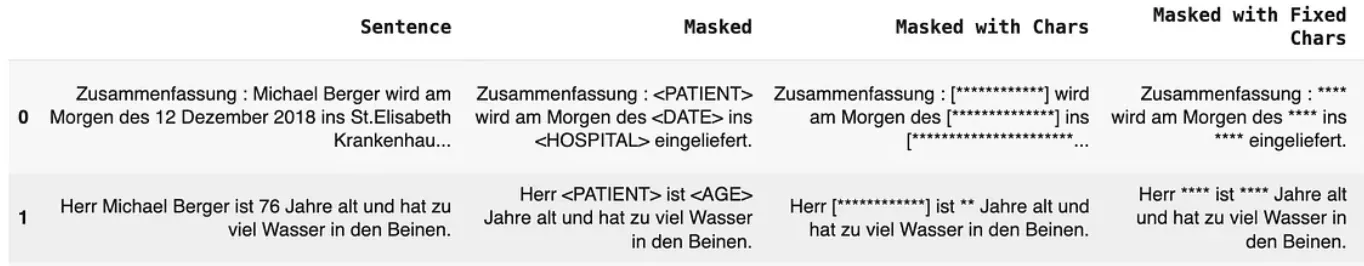
Masking
Masking involves replacing PHI with placeholder text to obscure sensitive information. Key masking strategies include:
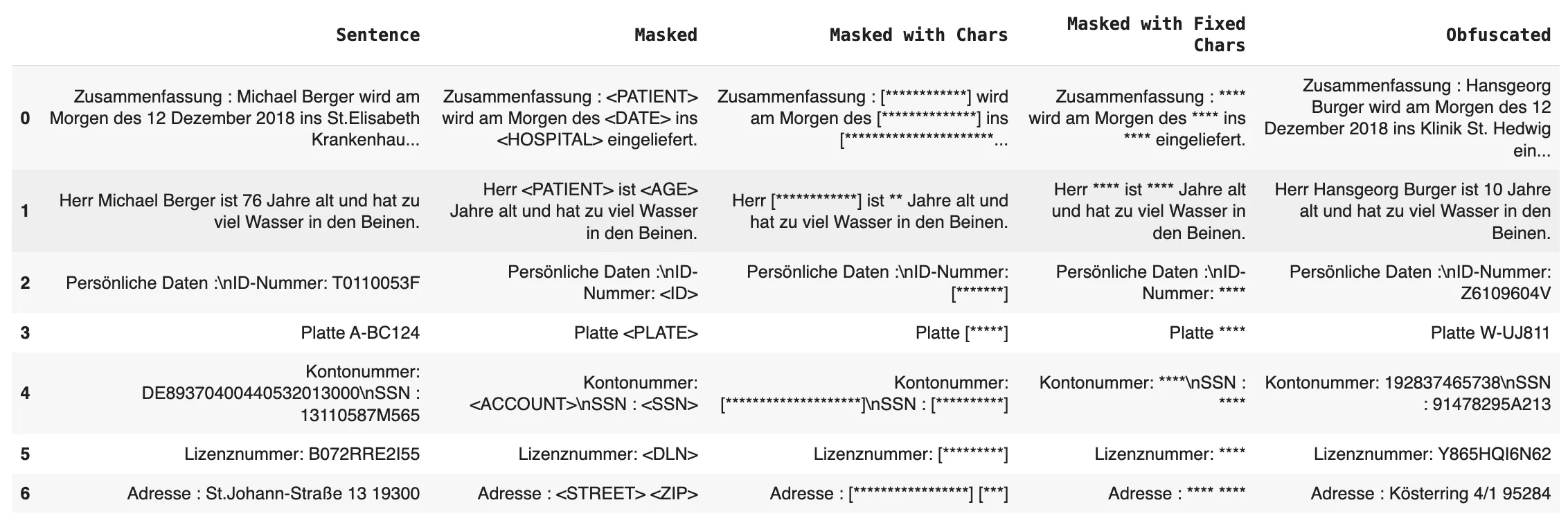
- Entity Labels: Replacing entities with their category label, e.g., [NAME] or [DATE].
- Same-Length Characters: Masking entities with the same number of asterisks, e.g., [***].
- Fixed-Length Characters: Replacing entities with a fixed number of asterisks, regardless of the entity’s original length.
Obfuscation
Obfuscation replaces sensitive entities with randomized but contextually appropriate values, ensuring the data remains realistic while anonymized. For example:
- Dates: Replacing a date like 03.2024 with a randomly generated date within a specified range.
- Names: Substituting names with randomized fake names.

De-Identification in Healthcare NLP: Using Models and Pipelines for Data Privacy
Implementing de-identification doesn’t have to be complex. Users can quickly get started by leveraging pretrained pipelines with just a few lines of code. These ready-to-use solutions ensure fast and efficient anonymization while maintaining the consistency and structure of the data.
This pipeline extract those labels: PATIENT , DOCTOR, HOSPITAL, DATE, ORGANIZATION, CITY, STREET, COUNTRY, USER NAME, PROFESSION, PHONE, AGE, CONTACT, ID, ZIP, ACCOUNT, SSN, DRIVER’S LICENSE NUMBER, PLATE NUMBER
from sparknlp.pretrained import PretrainedPipeline
deid_pipeline_de = PretrainedPipeline("clinical_deidentification", "de", "clinical/models")
For those with specific privacy requirements, Healthcare NLP allows for the creation of custom pipelines by combining different models to fit unique de-identification needs. Whether handling multilingual data, adapting to domain-specific terms, or applying advanced entity replacement strategies, our framework provides flexibility to ensure compliance with regulatory standards.
The Healthcare NLP Library offers two key models for de-identifying sensitive information in German medical texts.
- Generic De-identification Model: This model is designed to identify a broad range of protected health information (PHI). It detects 7 entities crucial for general de-identification tasks. Entities labeled by the Generic Model:
DATE,NAME,LOCATION,PROFESSION,AGE,ID,CONTACT - Subentity De-identification Model: This model provides a more granular approach by detecting 12 entities, focusing on detailed PHI in medical texts. Entities labeled by the Subentity Model:
PATIENT,HOSPITAL,DATE,ORGANIZATION,CITY,STREET,USERNAME,PROFESSION,PHONE,COUNTRY,DOCTOR,AGE
Let’s create our custom pipeline with these 2 de-idetification models.
Base Pipeline Stages:
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentence_detector = SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
word_embeddings = WordEmbeddingsModel.pretrained("w2v_cc_300d","de","clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("embeddings")
Two NER models at the same pipeline:
deid_ner = MedicalNerModel.pretrained("ner_deid_subentity", "de", "clinical/models")\
.setInputCols(["sentence", "token", "embeddings"])\
.setOutputCol("ner")
ner_converter = NerConverterInternal()\
.setInputCols(["sentence", "token", "ner"])\
.setOutputCol("ner_deid_subentity_chunk")
deid_ner_generic = MedicalNerModel.pretrained("ner_deid_generic", "de", "clinical/models")\
.setInputCols(["sentence", "token", "embeddings"])\
.setOutputCol("deid_ner_generic")
ner_converter_generic = NerConverterInternal()\
.setInputCols(["sentence", "token", "deid_ner_generic"])\
.setOutputCol("ner_deid_generic_chunk")
chunk_merger = ChunkMergeApproach()\
.setInputCols("ner_deid_subentity_chunk", 'ner_deid_generic_chunk')\
.setOutputCol('merged_ner_chunk')
De-identification stages:
deid_masked_entity = DeIdentification()\
.setInputCols(["sentence", "token", "merged_ner_chunk"])\
.setOutputCol("masked_with_entity")\
.setMode("mask")\
.setMaskingPolicy("entity_labels")\
deid_masked_char = DeIdentification()\
.setInputCols(["sentence", "token", "merged_ner_chunk"])\
.setOutputCol("masked_with_chars")\
.setMode("mask")\
.setMaskingPolicy("same_length_chars")\
deid_masked_fixed_char = DeIdentification()\
.setInputCols(["sentence", "token", "merged_ner_chunk"])\
.setOutputCol("masked_fixed_length_chars")\
.setMode("mask")\
.setMaskingPolicy("fixed_length_chars")\
.setFixedMaskLength(4)\
deid_obfuscated_faker = DeIdentification()\
.setInputCols(["sentence", "token", "merged_ner_chunk"]) \
.setOutputCol("obfuscated") \
.setMode("obfuscate")\
.setLanguage('de')\
.setObfuscateDate(True)\
.setObfuscateRefSource('faker')
nlpPipeline_de = Pipeline(
stages=[
document_assembler,
sentence_detector,
tokenizer,
word_embeddings,
deid_ner,
ner_converter,
deid_ner_generic,
ner_converter_generic,
chunk_merger,
deid_masked_entity,
deid_masked_char,
deid_masked_fixed_char,
deid_obfuscated_faker
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
model_de = nlpPipeline_de.fit(empty_data)
Let’s check the result:

To further improve the accuracy of entity recognition during the de-identification process, rule-based matchers can be integrated into the NER stages of the Healthcare NLP pipeline. These matchers complement the existing DL models by targeting specific patterns, improving detection of complex or edge-case entities that may be missed by machine learning models alone.
With these options, organizations can seamlessly integrate de-identification into their workflows, balancing privacy, usability, and compliance.
Conclusion
Protecting patient privacy is more than a legal obligation; it is a core responsibility in healthcare. The process of de-identification plays a vital role by enabling the secure use of sensitive medical data in research, diagnostics, and healthcare collaboration. It ensures that patient confidentiality is maintained while still allowing data to be repurposed for essential studies such as drug development, disease research, and public health analyses.
For organizations working with German medical texts, de-identification poses unique challenges due to complex language structures, specialized terminology, and the high density of sensitive information. However, the Healthcare NLP Library offers comprehensive solutions designed to address these issues. With over 2,500 pre-trained models, including both generic and subentity de-identification models, users can efficiently anonymize key entities such as names, dates, locations, patient identifiers, and more. These models have been tailored specifically to the needs of healthcare professionals and researchers handling German-language data, ensuring compliance with GDPR and other privacy regulations.
Key benefits of our de-identification solutions include:
- Local or Offline Processing: For maximum data security, organizations can deploy our models entirely offline in secure environments without the need for internet connectivity.
- Scalability: The system is capable of processing large datasets, allowing institutions to manage millions of clinical records with high performance and efficiency.
- Customization: In addition to our pre-built pipelines, the library allows users to train and customize models to meet specific organizational needs, such as handling rare or domain-specific entities.
- Ongoing Support: Users have access to dedicated support through Slack channels and can engage directly with experts and developers. Comprehensive certification training notebooks and tutorials are also available for rapid onboarding.
De-identification is a cornerstone of responsible data management in healthcare. By anonymizing data effectively, institutions can unlock new research opportunities, share information securely, and train advanced DL models — all while protecting the privacy of their patients.
Healthcare NLP models are licensed, so if you want to use these models, you can watch “Get a Free License For John Snow Labs NLP Libraries” video and request one license here.