In an era of rapidly advancing healthcare technology, the protection of patient privacy is more critical than ever. Medical records, rich with sensitive information, are invaluable for research and innovation but must be carefully managed to ensure compliance with regulations like HIPAA and GDPR. Deidentification, the process of removing or obscuring personally identifiable information (PII) from medical data, lies at the heart of this effort.
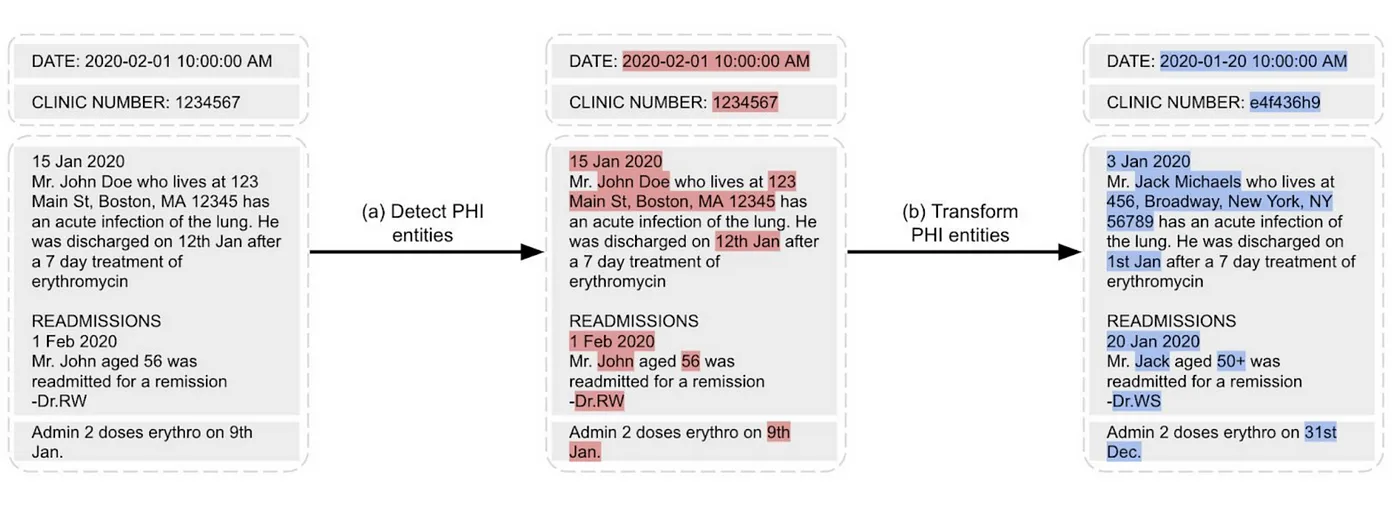
De-Identification process identifies potential pieces of content with personal information about patients and removes them by replacing them with semantic tags or fake entities.
Leveraging LLMs for de-identifying sensitive data (PHI) might be considered excessive and potentially unreliable, depending on the use case and the required level of customization. While LLMs like ChatGPT are highly capable of generating high-quality text, they are not specifically designed or optimized for the precise task of PHI de-identification.
This blog explores the performance and comparison of de-identification services provided by Healthcare NLP, Amazon, Azure, OpenAI, and Anthropic Claude focusing on their accuracy when applied to a dataset annotated by healthcare experts.
This comparison is valuable because it provides critical insights into the strengths and limitations of each de-identification solution, enabling researchers, developers, and organizations to make informed decisions when choosing a tool. For researchers, it helps identify the most accurate and reliable service for sensitive data processing. For developers, it highlights the ease of integration and API flexibility, which are crucial for building scalable solutions. For organizations, particularly in healthcare and finance, it offers a clear perspective on compliance capabilities, and performance, ensuring the selected tool meets regulatory requirements while optimizing operational efficiency.
Dataset
For this benchmark, we utilized 48 open-source documents annotated by domain experts from John Snow Labs. The annotations focused on the extraction of IDNUM, LOCATION, DATE, AGE, NAME, and CONTACT entities because these labels represent critical personal information that is commonly targeted for deidentification in various healthcare. These labels are typically associated with sensitive aspects of a person’s identity and are often required to be removed or anonymized in compliance with regulations like HIPAA or GDPR. By centering the benchmark around these entities, the task ensures that the performance evaluation is directly relevant to the challenges of real-world deidentification, which often involves identifying and obscuring such critical personal data.

Ground Truth Dataset
Tools Compared
Healthcare NLP & LLM
The Healthcare Library is a powerful component of John Snow Labs’ Spark NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,500 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs has created custom large language models (LLMs) tailored for diverse healthcare use cases. These models come in different sizes and quantization levels, designed to handle tasks such as summarizing medical notes, answering questions, performing retrieval-augmented generation (RAG), named entity recognition and facilitating healthcare-related chats.
John Snow Labs’ Healthcare NLP & LLM library offers a powerful solution to streamline the de-identification of medical records. By leveraging advanced Named Entity Recognition (NER) models, the library can automatically identify and deidentify Protected Health Information (PHI) from unstructured clinical notes. This capability ensures compliance with privacy regulations while maintaining the utility of the data for research and analysis. With this technology, healthcare organizations can efficiently anonymize sensitive information, enabling the safe sharing of clinical data for studies, improving patient privacy, and fostering innovation in medical research.
With the Healthcare NLP library, you can either build a custom de-identification pipeline to target specific labels or use pretrained pipelines with just two lines of code to deidentify a wide range of entities, including AGE, CONTACT, DATE, ID, LOCATION, NAME, PROFESSION, CITY, COUNTRY, DOCTOR, HOSPITAL, IDNUM, MEDICALRECORD, ORGANIZATION, PATIENT, PHONE, STREET, USERNAME, ZIP, ACCOUNT, LICENSE, VIN, SSN, DLN, PLATE, IPADDR, and EMAIL.
For this benchmark, we specifically used the clinical_de-identification_docwise_benchmark pretrained pipeline, which is designed to extract and de-identify NAME, IDNUM, CONTACT, LOCATION, AGE, DATE entities. It’s important to note that this pipeline does not rely on any LLM components.
Here is a sample code for using the Healthcare NLP pipeline:
from sparknlp.pretrained import PretrainedPipeline
deid_pipeline = PretrainedPipeline("clinical_deidentification_docwise_benchmark", "en", "clinical/models")
sample_text = "Patient John Doe was admitted to Boston General Hospital on 01/12/2023."
result = deid_pipeline.fullAnnotate(sample_text)
Azure Health Data Services
Azure Health Data Services’ de-identification service is designed to protect sensitive health information while preserving data utility. This API leverages natural language processing techniques to identify, label, redact, or surrogate Protected Health Information (PHI) in unstructured medical texts. Launched in 2024, the service offers three key operations: Tag, Redact, and Surrogate, enabling healthcare organizations to process diverse types of clinical documents securely and efficiently.
The de-identification service stands out for its ability to balance privacy concerns with data usability. By employing machine learning algorithms, it can detect HIPAA’s 18 identifiers and other PHI entities, ensuring compliance with various regional privacy regulations such as GDPR and CCPA. This innovative tool empowers healthcare professionals, researchers, and organizations to unlock the potential of their clinical data for machine learning, analytics, and collaborative research while maintaining the highest standards of patient privacy and data protection. You can get more details from here: https://learn.microsoft.com/en-us/azure/healthcare-apis/deidentification/
Here is a sample code for using Azure Health Data Services:
import os
CREDENTIALS_IN_JSON_FORMAT = {
"AZURE_CLIENT_ID":"", # update here with yours
"AZURE_TENANT_ID":"", # update here with yours
"AZURE_CLIENT_SECRET":"" # update here with yours
}
for k, v in CREDENTIALS_IN_JSON_FORMAT.items():
os.environ[k] = v
# This is constant for one app setup
os.environ['AZURE_HEALTH_DEIDENTIFICATION_ENDPOINT'] = '' # update here
from azure.health.deidentification import DeidentificationClient
from azure.identity import DefaultAzureCredential
from azure.health.deidentification.models import *
endpoint = os.environ["AZURE_HEALTH_DEIDENTIFICATION_ENDPOINT"]
endpoint = endpoint.replace("https://", "")
credential = DefaultAzureCredential(exclude_interactive_browser_credential=False)
client = DeidentificationClient(endpoint, credential)
sample_text = "Patient John Doe was admitted to Boston General Hospital on 01/12/2023."
body = DeidentificationContent(input_text=sample_text, operation="tag")
result: DeidentificationResult = client.deidentify(body)
Amazon Comprehend Medical
Amazon Comprehend Medical is a HIPAA-eligible natural language processing (NLP) service that leverages machine learning to extract valuable health data from unstructured medical text. This powerful tool can quickly and accurately identify medical entities such as conditions, medications, dosages, tests, treatments, and protected health information (PHI) from various clinical documents including physician’s notes, discharge summaries, and test results. With its ability to understand context and relationships between extracted information, Amazon Comprehend Medical offers a robust solution for healthcare professionals and researchers looking to automate data extraction, improve patient care, and streamline clinical workflows. You can get more details from here: https://docs.aws.amazon.com/comprehend-medical/latest/dev/textanalysis-phi.html
Here is a sample code for using Amazon Comprehend Medical:
import boto3
# Extract validated credentials for role assumption
validated_access_key_id = MFA_validated_token['Credentials']['AccessKeyId']
validated_secret_access_key = MFA_validated_token['Credentials']['SecretAccessKey']
validated_session_token = MFA_validated_token['Credentials']['SessionToken']
temp_sts_client = boto3.client(
'sts',
aws_access_key_id=validated_access_key_id,
aws_secret_access_key=validated_secret_access_key,
aws_session_token=validated_session_token
)
# choose your role
target_role_arn = "" # update here
# Assume the desired role
response = temp_sts_client.assume_role(
RoleArn=target_role_arn,
RoleSessionName='MedComp'
)
# temporary creds
tmp_access_key_id = response['Credentials']['AccessKeyId']
tmp_secret_access_key = response['Credentials']['SecretAccessKey']
tmp_session_token = response['Credentials']['SessionToken']
client = boto3.client(service_name='comprehendmedical',
region_name='', # update here
aws_access_key_id = tmp_access_key_id,
aws_secret_access_key = tmp_secret_access_key,
aws_session_token = tmp_session_token
)
sample_text = "Patient John Doe was admitted to Boston General Hospital on 01/12/2023."
result = client.detect_phi(Text= sample_text)
Open AI GPT-4o for Deidentification
GPT-4o is a multimodal model that brings faster response times and improved classification accuracy compared to GPT-4, potentially making it even better at identifying and redacting sensitive information through smart prompting. While GPT-3.5 and GPT-4 have been thoroughly tested for de-identification, especially in handling medical text, GPT-4o stands out as a promising new option thanks to its performance boost across various tasks.
However, there’s still no formal research evaluating GPT-4o’s de-identification abilities. Since protecting PHI (Protected Health Information) is critical in healthcare AI, it’s important to understand how well GPT-4o performs in this area. Until we see empirical studies directly assessing its capabilities, its effectiveness in de-identification remains an exciting, yet unproven, possibility.
import openai
# OpenAI API Configuration
openai.api_key = "your_api_key_here"
template = """
You are an expert medical annotator with extensive experience in labeling medical entities within clinical texts. Your role is to accurately identify and annotate Protected Health Information (PHI) entities in the provided text, following the specified entity types.
### Instructions:
1. **Review the Text**: Carefully read the text to understand its medical context.
2. **Identify PHI Entities**: Locate any terms or phrases that represent PHI, based on the following entity types:
- IDNUM, LOCATION, DATE, AGE, NAME, CONTACT
3. **Annotate Entities**: For each identified PHI, provide the start and end character indices, the entity type, and the exact text (chunk) of the entity.
4. **Response Format**: Return the annotations in a structured JSON format, as demonstrated in the examples below.
### Example:
**Input Sentence:**
"MD Connect Call 11:59pm 2/16/69 from Dr. Hale at Senior Care Clinic Queen Creek, SD regarding Terri Bird."
**Annotated Entities:**
[
{'begin': 24, 'end': 30, 'entity_type': 'DATE', 'chunk': '2/16/69'},
{'begin': 42, 'end': 45, 'entity_type': 'NAME', 'chunk': 'Hale'},
{'begin': 50, 'end': 67, 'entity_type': 'LOCATION', 'chunk': 'Senior Care Clinic'},
{'begin': 69, 'end': 79, 'entity_type': 'LOCATION', 'chunk': 'Queen Creek'},
{'begin': 83, 'end': 84, 'entity_type': 'LOCATION', 'chunk': 'SD'},
{'begin': 96, 'end': 105, 'entity_type': 'NAME', 'chunk': 'Terri Bird'}
]
---
### Task:
Extract all PHI entities from the text below. The entity types to identify are: **IDNUM, LOCATION, DATE, AGE, NAME, CONTACT**.
**Expected Output Format:**
{
'entities':[
{'begin': , 'end': , 'entity_type': '', 'chunk': ''}
]
}
---
### Text to Annotate:
{text}
---
### Your Response:
"""
def get_llm_inference(text):
try:
# Create the prompt by inserting the text into the template
prompt = template.format(text=text)
# Make API call to OpenAI
response = openai.Completion.create(
model="gpt-4o", # Use the GPT-4o model
prompt=prompt,
temperature=0.1,
max_tokens=1500, # Adjust as needed for long texts
stop=["###", "\n"] # Ensure the response ends cleanly
)
# Parse the response to extract the entities
llm_output = response.choices[0].text.strip()
# Return the entities from the output (assumes the output is valid JSON)
return llm_output # You can use json.loads to parse if necessary
except Exception as e:
print(f"ERROR: {e}")
return None
# Example usage
sample_text = "Patient John Doe was admitted to Boston General Hospital on 01/12/2023."
result = get_llm_inference(sample_text)
print(result)
Anthropic Claude 3.7 Sonnet
Claude 3.7 Sonnet, a mid-tier model in Anthropic’s Claude 3 family, balances speed, efficiency, and advanced language understanding. Its ability to handle complex reasoning while maintaining high accuracy in text processing makes it particularly well-suited for healthcare AI applications. When applied to PHI extraction, the model demonstrates strong contextual awareness, allowing it to identify and categorize sensitive patient data within medical notes. With built-in safety features and scalability, Claude 3.7 Sonnet presents a promising solution for streamlining de-identification workflows in both clinical and research settings.
However, no formal research has yet assessed Claude 3.7 Sonnet’s de-identification capabilities. Given the importance of protecting Protected Health Information (PHI) in healthcare AI, evaluating its effectiveness in this area is essential.
prompt = f"""
You are an expert medical annotator with extensive experience in labeling medical entities within clinical texts. Your role is to accurately identify and annotate Protected Health Information (PHI) entities in the provided text, following the specified entity types.
### Instructions:
1. **Review the Text**: Carefully read the text to understand its medical context.
2. **Identify PHI Entities**: Locate any terms or phrases that represent PHI, based on the following entity types:
- IDNUM, LOCATION, DATE, AGE, NAME, CONTACT
3. **Annotate Entities**: For each identified PHI, provide the start and end character indices, the entity type, and the exact text (chunk) of the entity.
4. **Response Format**: Return the annotations in a structured JSON format, as demonstrated in the examples below.
5. DO NOT return any other text or explanation like 'Here is the entities..' or 'JSON:...'.
### Example:
**Input Sentence:**
"MD Connect Call 11:59pm 2/16/69 from Dr. Hale at Senior Care Clinic Queen Creek, SD regarding Terri Bird."
**Annotated Entities:**
[
{{'begin': 24, 'end': 30, 'entity_type': 'DATE', 'chunk': '2/16/69'}},
{{'begin': 42, 'end': 45, 'entity_type': 'NAME', 'chunk': 'Hale'}},
{{'begin': 50, 'end': 67, 'entity_type': 'LOCATION', 'chunk': 'Senior Care Clinic'}},
{{'begin': 69, 'end': 79, 'entity_type': 'LOCATION', 'chunk': 'Queen Creek'}},
{{'begin': 83, 'end': 84, 'entity_type': 'LOCATION', 'chunk': 'SD'}},
{{'begin': 96, 'end': 105, 'entity_type': 'NAME', 'chunk': 'Terri Bird'}}
]
### Task:
Extract all PHI entities from the text below. DO NOT return any other text or explanation like 'Here is the entities..'. The ONLY PHI entity types to identify are: **IDNUM, LOCATION, DATE, AGE, NAME, CONTACT**.
Expected Output Format:
{{
entities:[
{{'begin': , 'end': , 'entity_type': '', 'chunk': ''}}
]
}}
### Text to Annotate:
{text}
### Your Response:
"""
import anthropic
# Initialize the Anthropic client
client = anthropic.Anthropic(api_key="") # Claude API key
def get_prediction(text):
prompt = get_prompt(text)
# Call Claude 3.7 Sonnet API
response = client.messages.create(
model="claude-3-7-sonnet-20250219", # Specify Claude 3.7 Sonnet model
max_tokens=5000, # Adjust as needed
messages=[{"role": "user", "content": prompt}]
)
return response
sample_text = "Patient John Doe was admitted to Boston General Hospital on 01/12/2023."
result = get_prediction(text)
Comparison of the Tools
The biggest difference between these tools comes down to flexibility. Azure Health Data Services, Amazon Comprehend Medical, GPT-4o, and Claude 3.7 Sonnet are API-based, black-box cloud solutions, meaning you can’t easily tweak or customize their outputs to fit specific needs. In contrast, the Healthcare NLP library offers a highly adaptable de-identification pipeline that you can run with just two lines of code. Not only can you customize the pipeline’s stages to suit your requirements, but you can also run it locally without needing an internet connection, giving you full control over your data processing.
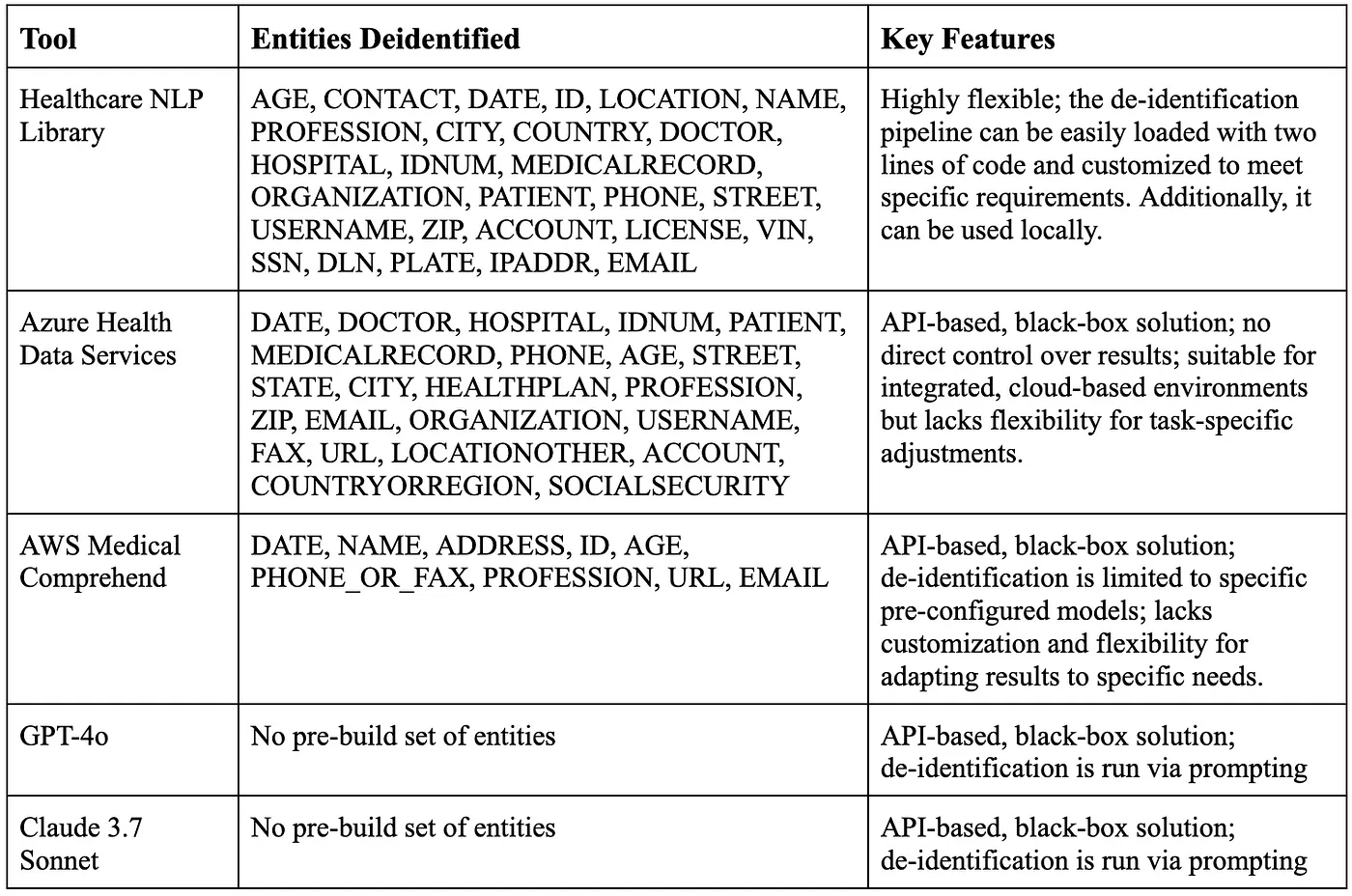
Comparison Table Of The Tools
You can find the code for using all these tools in the Deidentification Performance Comparison Of Healthcare NLP VS Cloud Solutions Notebook.
Evaluation Criteria
In this benchmark study, we employed two distinct approaches to compare accuracy:
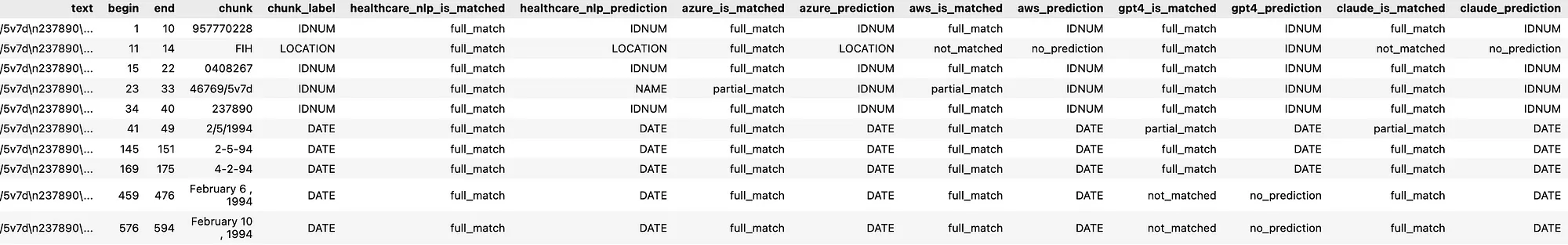
A. Entity-Level Evaluation: Since de-identifying PHI data is a critical task, we evaluated how well de-identification tools detected entities present in the annotated dataset, regardless of their specific labels in the ground truth. The detection outcomes were categorized as:
- full_match: The entire entity was correctly detected.
- partial_match: Only a portion of the entity was detected.
- not_matched: The entity was not detected at all.
For example:
Text: “Patient John Doe was admitted to Boston General Hospital on 01/12/2023.”
Ground Truth Entity: John Doe (NAME)
Predicted Entity: John Doe (NAME) ==> full_match
Predicted Entity: John==> partial_match
If predictions don’t have any match with “John Doe”, the result is “not_matched”.
B. Token-Level Accuracy: The text in the annotated dataset was tokenized, and the ground truth labels assigned to each token were compared with predictions made by the Healthcare NLP library, Amazon Comprehend Medical, Azure Health Data Services, GPT-4o and Claude 3.7 Sonnet models. Classification reports were generated for each tool, comparing their precision, recall, and F1 scores.
This dual approach comprehensively evaluated each tool’s performance in de-identifying PHI data.
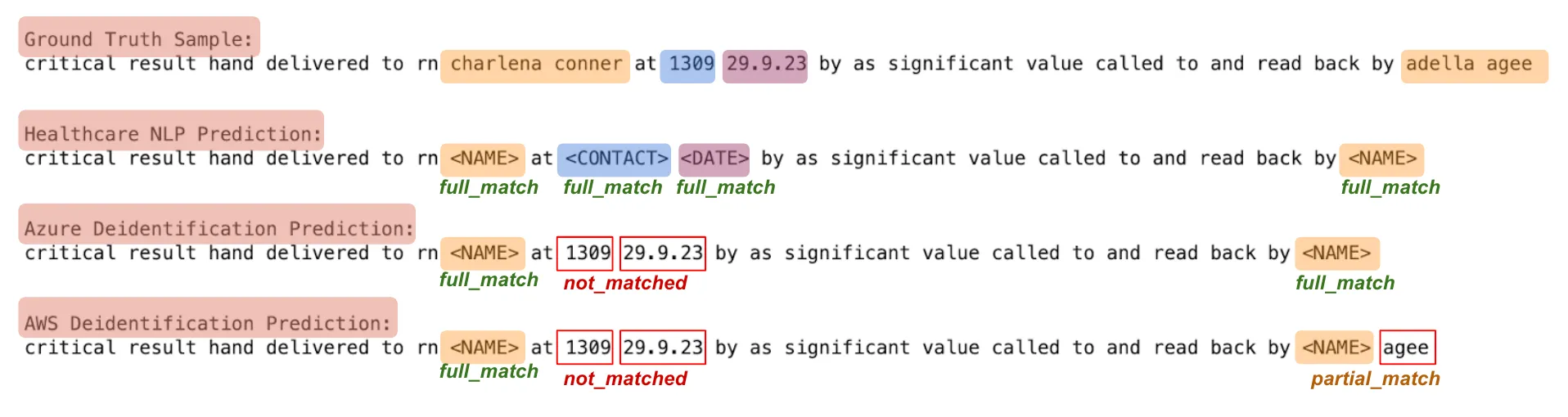
De-identification Results of the Tools on a Sample Text
Methodology
In this study, some differences were observed between the predictions of the de-identification services and the ground truth labels. In the ground truth dataset, entities were annotated with more generic labels. For example, all names were annotated as NAME instead of using labels like PATIENT_NAME for patient names and DOCTOR_NAME for doctor names. To ensure consistency, the labels from the de-identification tools’ predictions were mapped to the corresponding ground truth labels.
Additionally, for a fair comparison, entities that could not be mapped to the ground truth labels (e.g., PROFESSION, ORGANIZATION, etc.) were excluded from the predictions before comparing the results.
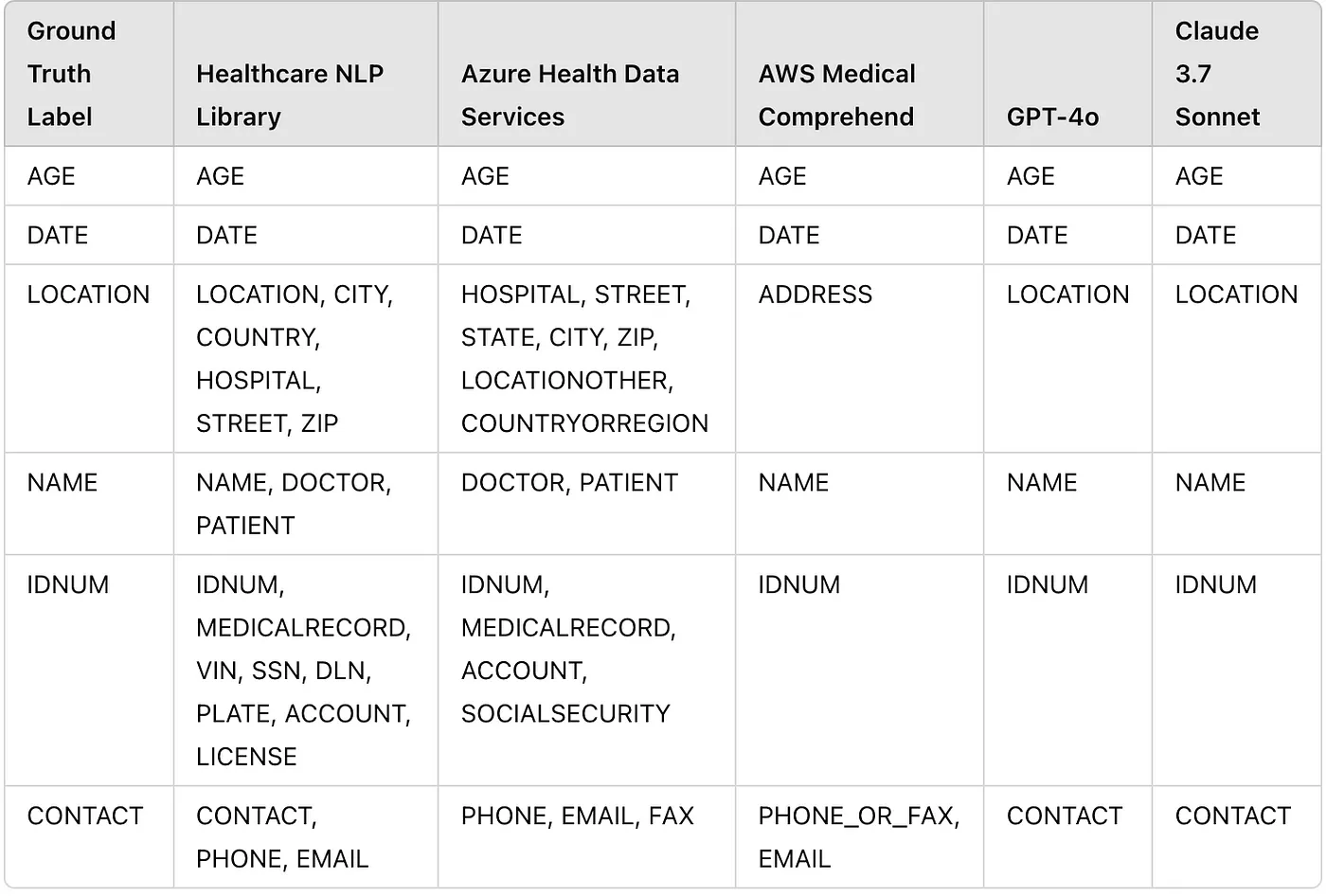
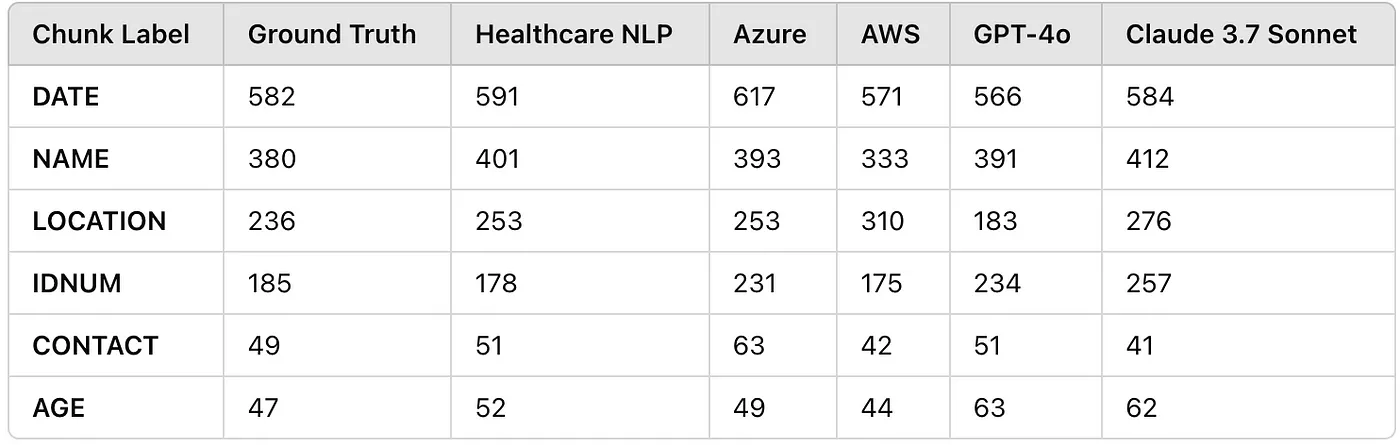
Entity Counts Table
Results and Findings
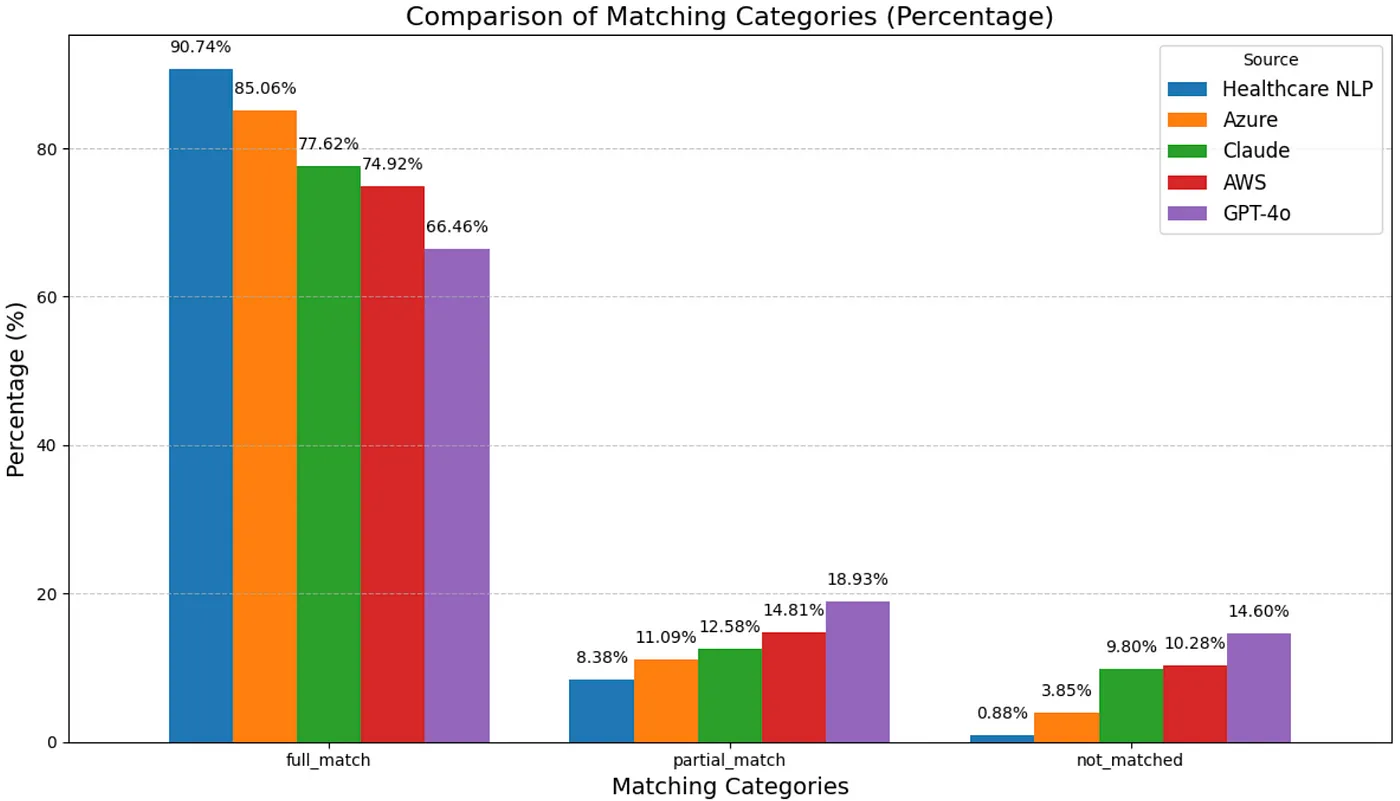
Entity-Level Evaluation
The results obtained from matching the predictions made by Healthcare NLP, Amazon Comprehend Medical, Azure Health Data Services, GPT-4o, and Claude 3.7 Sonnet with the ground truth entities are as follows:
Token-Level Evaluation
The resulting data frame, obtained by tokenizing the text annotated as ground truth and adding the corresponding ground truth, Healthcare NLP, Amazon Comprehend Medical, Azure Health Data Services, GPT-4o, and Claude 3.7 Sonnet prediction labels for each token, is as follows:
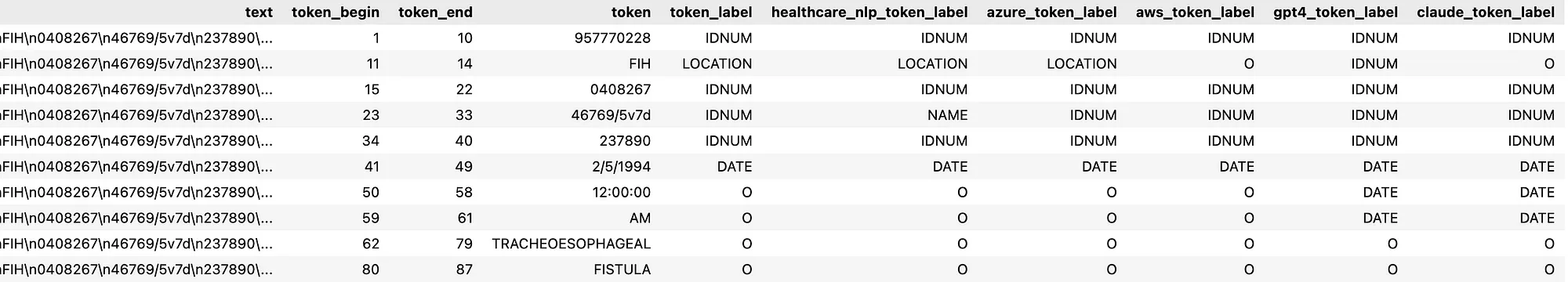
Token Level Results Dataframe
Based on the token-level results, the classification reports showing the comparison between each de-identification tool’s predictions and the ground truth labels are as follows:
Healthcare NLP:
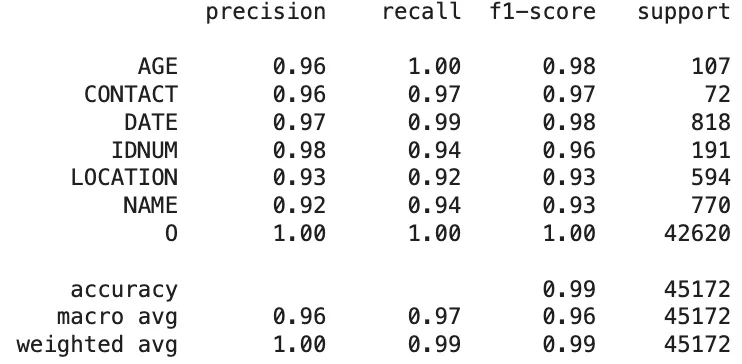
Classification Report of The Healthcare NLP Library Predictions
Azure Health Data Services:
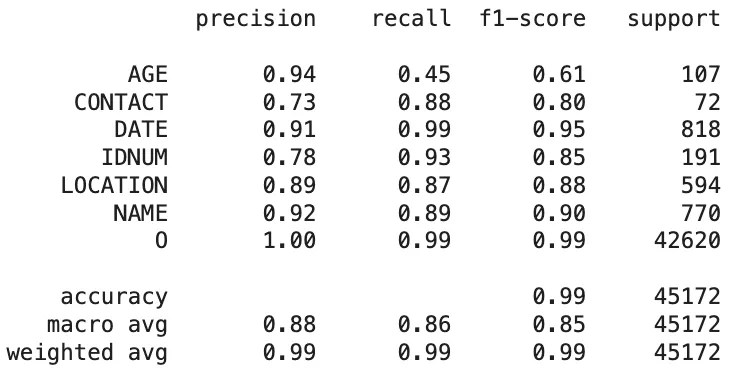
Classification Report of The Azure Health Data Services Predictions
Amazon Comprehend Medical:
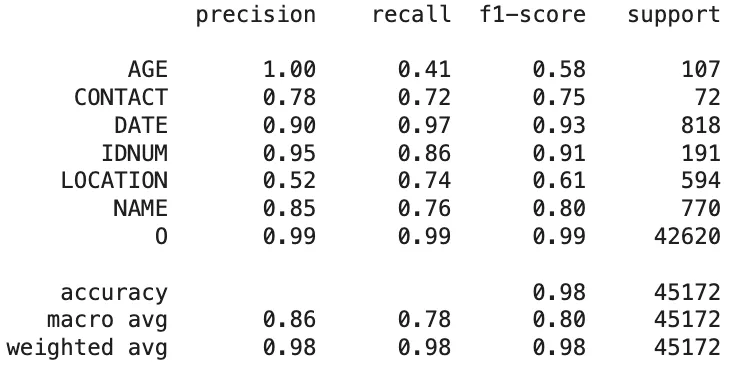
Classification Report of The Amazon Comprehend Medical Predictions
OpenAI GPT-4o Model:
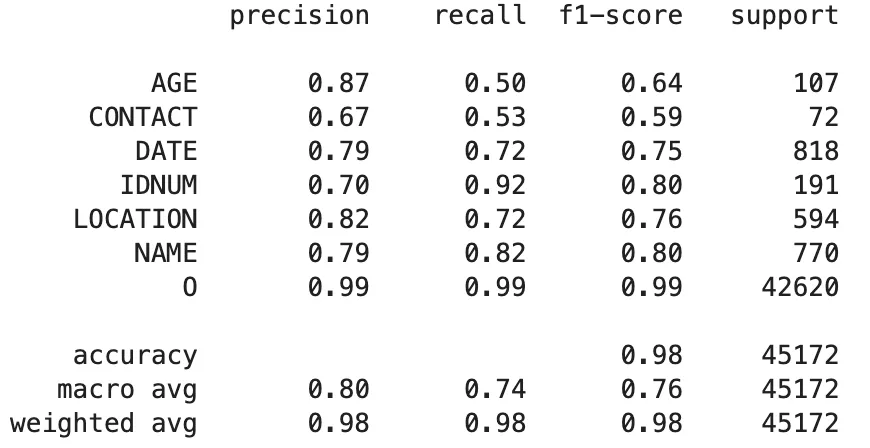
Classification Report of The GPT-4o Model Predictions
Anthropic Claude 3.7 Sonnet:
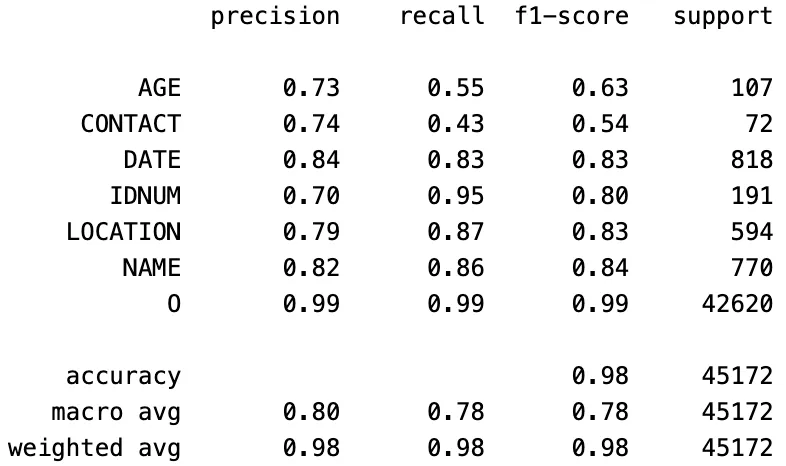
Classification Report of The Claude 3.7 Sonnet Model Predictions
Let’s visualize the F1-Scores for each label on a single plot:
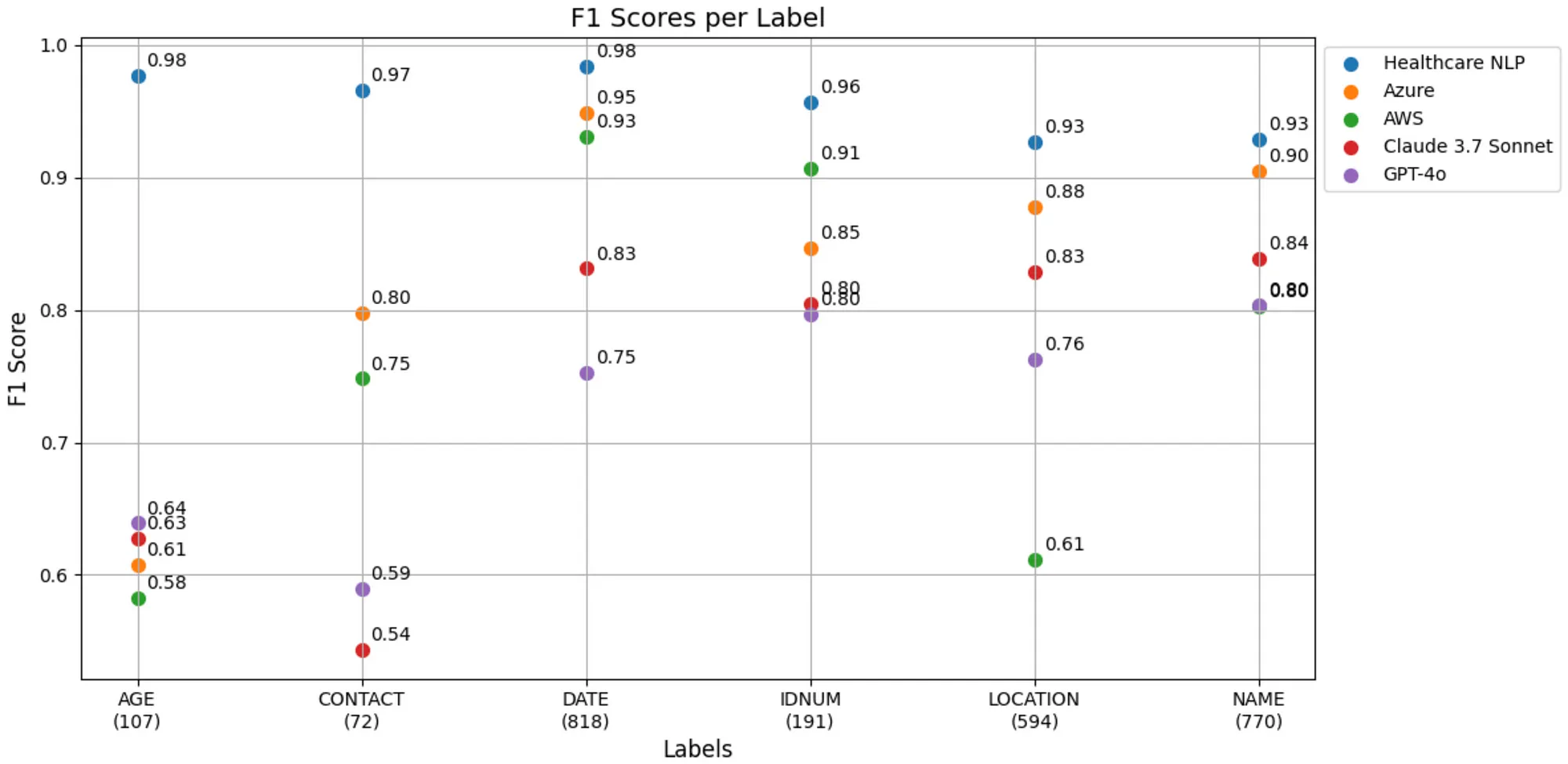
F1-Score Table for Each Label
PHI Entity Prediction Comparison
The de-identification task is crucial in ensuring the privacy of sensitive data. In this context, the primary focus is not on the specific labels of PHI entities but on whether they are successfully detected. When evaluating the results based on the classification of entities as PHI or non-PHI, the outcomes for PHI entity detection are as follows:
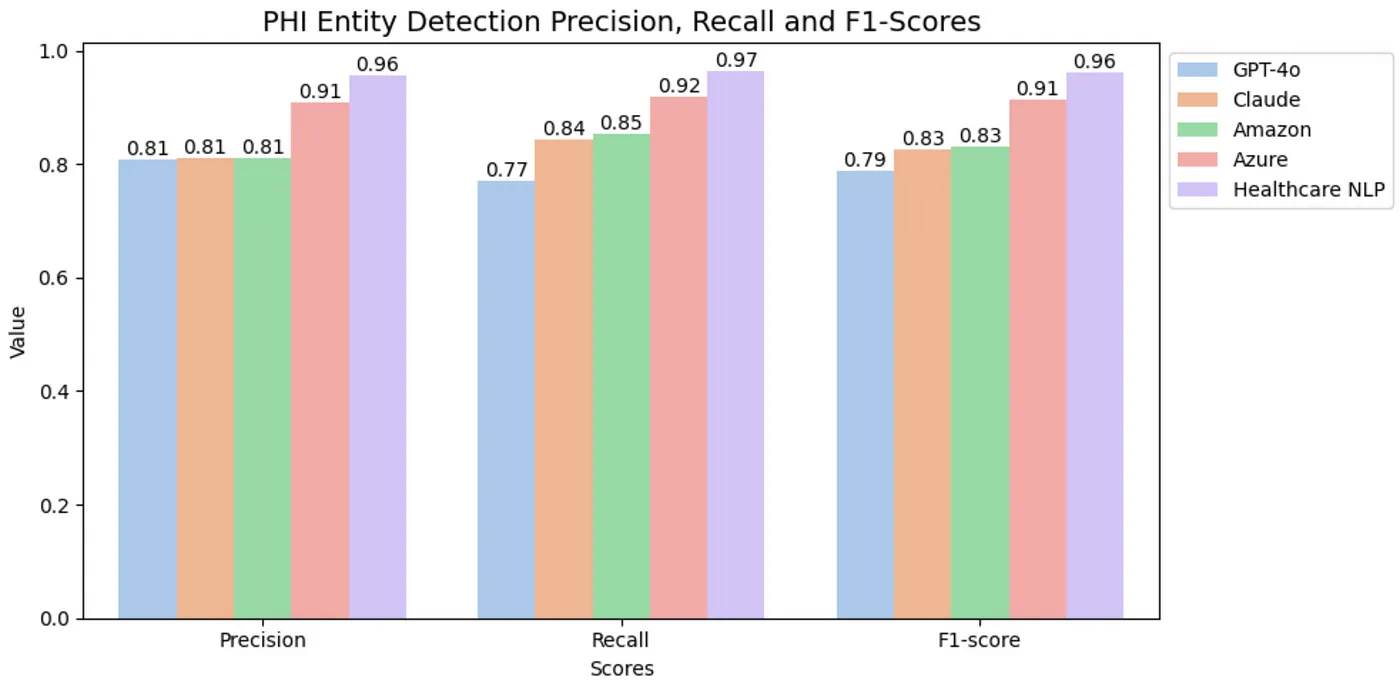
PHI Entity Detection Scores
Price Analysis of the Tools
When handling large volumes of clinical notes, cost becomes a key consideration. Since such small datasets are rare in real-world applications, we estimated the expenses for these tools based on processing 1 million unstructured clinical notes, with each document averaging around 5,250 characters.
Azure Health Data Services Pricing: Based on the pricing page, generating PHI predictions for 1M documents, each averaging 5,250 characters, costs $13,125.
Amazon Comprehend Medical Pricing: According to the price calculator, obtaining PHI predictions for 1M documents, with an average of 5,250 characters per document, costs $14,525.
Open AI GPT-4o Pricing: Processing 1M documents costs approximately $21,400.
Anthropic Claude 3.7 Sonnet: Processing 1M documents costs approximately $23,330.
Healthcare NLP Pricing: When using John Snow Labs-Healthcare NLP Prepaid product on an EC2–32 CPU (c6a.8xlarge at $1,2 per hour) machine, obtaining the de-identification codes for PHI entities from approximately 48 documents takes around 39.4 seconds. Based on this, processing 1M documents and de-identifying the PHI entities would take about 13680 minutes (228 hours (approximately 9.5 days), but with proper scaling, it can be completed in just one day), costing $273 for infrastructure and $2145 for the license (considering a 1-month license price of $7,000). Thus, the total cost for Healthcare NLP is approximately $2418.
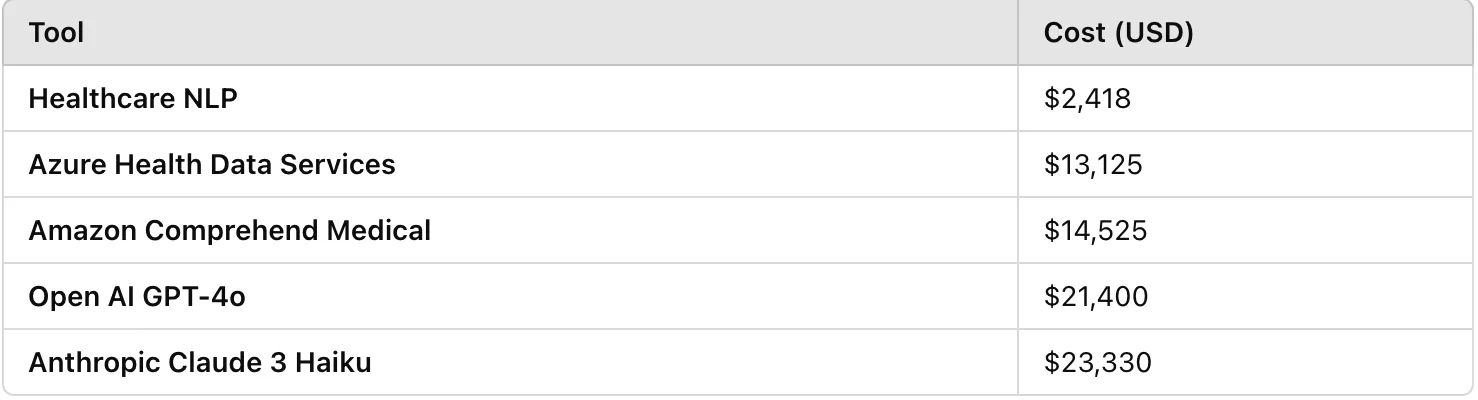
Cost Comparison of Tools for Processing 1 Million Clinical Notes
Based on our estimates for processing 1 million clinical notes, Azure, Amazon, GPT-4o, and Claude come with significantly higher costs, whereas Healthcare NLP allows organizations to perform de-identification at a fraction of the price. This makes it an ideal choice for institutions handling large volumes of data, providing both affordability and customization without compromising performance.
Conclusion
In this study, we compared the performance of Healthcare NLP, Amazon Comprehend Medical, Azure Health Data Services, OpenAI GPT-4o, and Anthropic Claude 3.7 Sonnet tools on a ground truth dataset annotated by medical experts. The results were evaluated from two perspectives: chunk-level and token-level.
- The chunk-level evaluation showed that Healthcare NLP outperformed the others in terms of accurately capturing entities and minimizing missed entities. After Healthcare NLP, Azure Health Data Services ranked second, followed by Claude 3.7 Sonnet model in third, Amazon Comprehend Medical in fourth, and GPT-4o model in fifth.
- The token-level evaluation revealed that Healthcare NLP achieved the best results in terms of precision, recall, and F1 score, with Azure Health Data Services, Amazon Comprehend Medical, Claude 3.7 Sonnet, and GPT-4o trailing behind in that order.
- Adaptability of Tools: One of the key differences among these tools is their level of adaptability. While Azure Health Data Services, Amazon Comprehend Medical, OpenAI GPT-4o, and Anthropic Claude 3.7 Sonnet model are API-based, black-box cloud solutions that do not allow modifications or customization of results, the Healthcare NLP library offers a highly flexible approach. Its de-identification pipeline can be implemented with just two lines of code, and users can easily customize outputs by modifying the pipeline’s stages to suit specific requirements.
- Cost-Effectiveness: Healthcare NLP stands out as the most cost-effective solution for de-identification when comparing tools (excluding Azure, as its pricing could not be verified) for processing large-scale clinical notes. In contrast to API-based services like Azure, Amazon, OpenAI, and Anthropic Claude models, which charge per request and can become costly at scale, Healthcare NLP provides a flexible, local deployment option that drastically reduces costs. Furthermore, regardless of processing 1 billion clinical notes, the cost remains fixed for the same time period. Unlike API-based solutions, which incur higher costs with increased volume, Healthcare NLP offers consistent and predictable pricing.
In summary, the Healthcare NLP library consistently outperformed Azure Health Data Services, Amazon Comprehend Medical, OpenAI GPT-4o , and Anthropic Claude 3.7 Sonnet model in both chunk-level and token-level evaluations, achieving the highest precision, recall, and F1 scores while minimizing missed entities. Beyond accuracy, its adaptability sets it apart, unlike Azure, Amazon, GPT-4o, and Claude 3.7 Sonnet which are black-box API solutions with no customization options, Healthcare NLP allows users to modify its deidentification pipeline to meet specific needs. Additionally, it proves to be the most cost-effective solution, offering substantial savings compared to cloud-based alternatives that charge per request.
You can find the whole code in this notebook: Deidentification Performance Comparison Of Healthcare NLP VS Cloud Solutions Notebook
Beyond its standalone capabilities, the Healthcare NLP library is also available on AWS, Snowflake, and Databricks Marketplaces, making it easier for organizations to integrate de-identification solutions into their existing workflows.
- Seamless Integration: Deploy directly within your preferred cloud ecosystem without a complex setup.
- Scalability: Process large-scale clinical data efficiently.
- Compliance & Security: Designed for healthcare-grade data protection, meeting HIPAA and GDPR standards.
Healthcare NLP models are licensed, so if you want to use these models, you can watch “Get a Free License For John Snow Labs NLP Libraries” video and request one license here.