Connect
Ontologies and extracted facts to form a knowledge graph
Discover
Visualize and explore the graph, or answer natural language questions
Biomedical Q&A at Roche
- The end-to-end system includes a user interface for graph visualization, exploration, search, and question answering
- Knowledge graph built at scale, combining multiple free-text data sources and ontologies
- Supports adding private documents and facts to the graph
- Tested on BioASQ challenge
Knowledge Graph
- 10k+ Predicates (Relation types)
- 500k Clinical Trials
- 85M Entities
- 260M Extracted Facts from Text
- 5B+ Total Facts
The Clinical Knowledge Graph
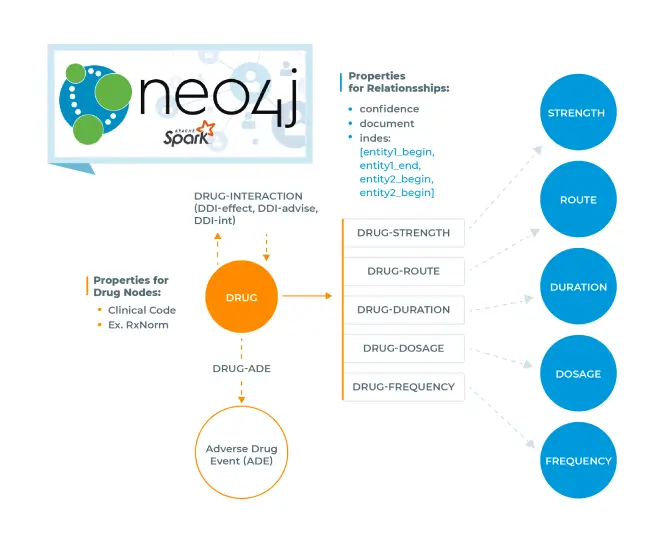
Combining Spark NLP & Neo4j
- Extracted named entities & relations between from
- Typical questions:
- What are the drug interactions and adverse drug events of “Lipitor”?
- What are the routes by which the drug containing API “Rofecoxib” is administered?
Automated question answering about clinical guidelines
- Joint work with Kaiser Permanente
- Answering both specific and general questions (‘how to treat diabetes?’)
- Automatic identification documents or section of documents with corresponding clinical guidelines – from curated set of clinical guidelines
- Indexing free text documents including PDF and DOCX
- Scalable to a large number of documents.
























