Medicine’s fundamental goal is to assist patients in managing and improving their health conditions. However, the language used within medical settings can sometimes inadvertently impact patients negatively, influencing their experiences and self-perception. Stigmatizing language in medical records can reduce patients to their medical conditions alone or cast judgments based on their illnesses, leading to diminished quality of care and exacerbated health inequalities. This article explores how stigmatizing language in medicine can be identified and addressed using a model developed by John Snow Labs.
The Impact of Stigmatizing Language in Medical Records
Language is a powerful tool that shapes perceptions, interactions, and outcomes. In healthcare, the words chosen by medical professionals can significantly influence patient-provider relationships and the overall treatment experience. Stigmatizing language in medical records not only affects how healthcare providers perceive and interact with patients but also impacts patients’ self-esteem, trust in medical systems, and willingness to seek care.
For instance, descriptors such as “non-compliant,” “aggressive,” or “resistant” can carry negative connotations, leading healthcare providers to develop unconscious biases toward patients. These biases may result in less empathetic care, misdiagnoses, or inadequate treatment plans. Patients who encounter or become aware of such language may feel devalued, ashamed, or reluctant to fully engage in their healthcare, further hindering their health outcomes.
Addressing stigmatizing language is therefore essential to promote equitable, compassionate, and effective healthcare. By recognizing and modifying such language, medical professionals can foster more respectful and supportive environments that encourage better patient engagement and health outcomes.
Also identifying stigmatizing language in clinical notes is crucial for several reasons:
- Compliance: With patients now having access to their medical records, ensuring that the language used is respectful and non-discriminatory is essential for legal and ethical compliance[1][2].
- Healthcare Equity: Stigmatizing language can exacerbate health disparities among marginalized groups. By addressing this issue, healthcare providers can promote equity and improve patient outcomes[1][2].
- Research on Bias: Understanding and mitigating bias in clinical documentation can enhance the quality of healthcare research and practice, ensuring that all patient populations are treated fairly[1][2].
Let’s start with a brief introduction to Spark NLP and then discuss the details of the Stigmatization NER model with some concrete results.
Healthcare NLP & LLM
The Healthcare Library is a powerful component of John Snow Labs’ Spark NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,200 + pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
At John Snow Labs, we have developed specialized LLM models of various sizes and quantification levels for many Healthcare applications (medical note summarization, enhancing diagnostic accuracy, Q&A, RAG and Chat).
John Snow Labs’ GitHub repository is a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to enhance further their understanding and utilization of Spark NLP and related tools.
John Snow Labs also offers periodic certification training to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the library’s capabilities. It allows users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
John Snow Labs and the Detection of Stigmatizing Language
Recognizing the profound impact of language in healthcare, John Snow Labs has developed an innovative model aimed at systematically analyzing and identifying stigmatizing language within medical records. This model leverages advanced natural language processing (NLP) techniques to detect and highlight language that may perpetuate stigma, allowing healthcare organizations to take corrective measures and promote more patient-centered communication.
To this end, we at JSL (John Snow Labs) categorized stigmatizing language as positive and negative and then trained a ner_stigmatization model using the labels below.
So how can we use this model?
For this, we need to set up a spark pipeline:
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentence_detector = SentenceDetectorDLModel.pretrained("sentence_detector_dl", "en")\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
clinical_embeddings = WordEmbeddingsModel.pretrained('embeddings_clinical', "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("embeddings")
ner_model = MedicalNerModel.pretrained("ner_stigmatization_wip", "en", "clinical/models")\
.setInputCols(["sentence", "token","embeddings"])\
.setOutputCol("ner")
ner_converter = NerConverterInternal()\
.setInputCols(['sentence', 'token', 'ner'])\
.setOutputCol('ner_chunk')
pipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
clinical_embeddings,
ner_model,
ner_converter
])
sample_texts = ["""During his hospital stay, David Brown's reluctance to seek care and resistance to necessary treatments highlighted the significant impact of poor reasoning and judgment on his health outcomes. His confrontational attitude and frequent defensiveness during discussions about his treatment plan revealed the deep-seated anxieties he harbored about his health. Despite these challenges, the healthcare team made concerted efforts to educate him on the importance of adhering to his prescribed regimen and attending regular follow-up appointments. However, Mr. Brown often fixated on incorrect beliefs, insisting that his symptoms were solely due to stress, which further complicated his care."""]
data = spark.createDataFrame(sample_texts, StringType()).toDF("text")
result = pipeline.fit(data).transform(data)
By systematically extracting and categorizing this information, NER models help build a structured dataset that forms the foundation for further analysis.
Extracting entities in a structured format improves usability and integration, enabling efficient retrieval and comprehensive analysis of patient information. It enhances consistency and standardization, supporting advanced analytical techniques and better decision-making. Overall, it transforms raw data into actionable insights, leading to improved patient care, effective research, and informed public health strategies.
PipelineTracer is a flexible class that tracks every stage of a pipeline. It provides detailed information about entities, assertions, de-identification, classification and relationships. This class also helps to build parser dictionaries to create a PipelineOutputParser. Some of the central functionality includes printing the pipeline schema, creating parser dictionaries, and retrieving possible assertions, relationships, and entities. Provide easy access to parser dictionaries and existing pipeline diagrams.
from sparknlp_jsl.pipeline_output_parser import PipelineOutputParser
light_model = LightPipeline(pipeline.fit(data))
light_result = light_model.fullAnnotate(sample_texts)
column_maps = {
'document_identifier': 'ner_clinical_pipeline',
'document_text': 'document',
'entities': ['ner_chunk'],
'assertions': [],
'resolutions': [],
'relations': [],
'summaries': [],
'deidentifications': [],
'classifications': []
}
pipeline_parser = PipelineOutputParser(column_maps)
result = pipeline_parser.run(light_result)
result['result'][0]
Result:
{'document_identifier': 'ner_clinical_pipeline',
'document_id': 0,
'document_text': ["During his hospital stay, David Brown's reluctance to seek care and resistance to necessary treatments highlighted the significant impact of poor reasoning and judgment on his health outcomes. His confrontational attitude and frequent defensiveness during discussions about his treatment plan revealed the deep-seated anxieties he harbored about his health. Despite these challenges, the healthcare team made concerted efforts to educate him on the importance of adhering to his prescribed regimen and attending regular follow-up appointments. However, Mr. Brown often fixated on incorrect beliefs, insisting that his symptoms were solely due to stress, which further complicated his care."],
'entities': [{'chunk_id': '285609be',
'chunk': 'reluctance',
'begin': 40,
'end': 49,
'ner_label': 'Resistant',
'ner_source': 'ner_chunk',
'ner_confidence': '0.992'},
{'chunk_id': '084d1318',
'chunk': 'resistance',
'begin': 68,
'end': 77,
'ner_label': 'Resistant',
'ner_source': 'ner_chunk',
'ner_confidence': '0.9999'},
{'chunk_id': '4b9266c2',
'chunk': 'treatments',
'begin': 92,
'end': 101,
'ner_label': 'TREATMENT',
'ner_source': 'ner_chunk',
'ner_confidence': '0.9952'},
{'chunk_id': 'ca710122',
'chunk': 'poor reasoning and judgment',
'begin': 141,
'end': 167,
'ner_label': 'Poor_Reasoning',
'ner_source': 'ner_chunk',
'ner_confidence': '0.99652493'},
{'chunk_id': '1feb4c15',
'chunk': 'confrontational',
'begin': 197,
'end': 211,
'ner_label': 'Argumentative',
'ner_source': 'ner_chunk',
'ner_confidence': '1.0'},
{'chunk_id': '343f80e7',
'chunk': 'defensiveness',
'begin': 235,
'end': 247,
'ner_label': 'Argumentative',
'ner_source': 'ner_chunk',
'ner_confidence': '1.0'},
{'chunk_id': '26a7fc0f',
'chunk': 'the deep-seated anxieties',
'begin': 302,
'end': 326,
'ner_label': 'PROBLEM',
'ner_source': 'ner_chunk',
'ner_confidence': '0.71680003'},
{'chunk_id': 'e1836c30',
'chunk': 'adhering',
'begin': 463,
'end': 470,
'ner_label': 'Compliant',
'ner_source': 'ner_chunk',
'ner_confidence': '1.0'},
{'chunk_id': '3dfb7add',
'chunk': 'his prescribed regimen',
'begin': 475,
'end': 496,
'ner_label': 'TREATMENT',
'ner_source': 'ner_chunk',
'ner_confidence': '0.8404667'},
{'chunk_id': 'bcb643cd',
'chunk': 'insisting',
'begin': 599,
'end': 607,
'ner_label': 'Credibility_Doubts',
'ner_source': 'ner_chunk',
'ner_confidence': '0.9986'},
{'chunk_id': 'db839d82',
'chunk': 'his symptoms',
'begin': 614,
'end': 625,
'ner_label': 'PROBLEM',
'ner_source': 'ner_chunk',
'ner_confidence': '0.97679996'},
{'chunk_id': 'c6966e75',
'chunk': 'stress',
'begin': 646,
'end': 651,
'ner_label': 'PROBLEM',
'ner_source': 'ner_chunk',
'ner_confidence': '0.9826'}],
'assertions': [],
'resolutions': [],
'relations': [],
'summaries': [],
'deidentifications': [],
'classifications': []}
John Snow Labs’ NER Visualizer provides a user-friendly interface for visualizing the results of NER models. It highlights and categorizes identified entities within the text. This tool allows users to see how the NER models extract and label entities, making it easier to understand and interpret the extracted data. The visualizer helps in validating the accuracy of the models, identifying patterns, and gaining insights from unstructured medical data, ultimately facilitating better data analysis and decision-making in healthcare.
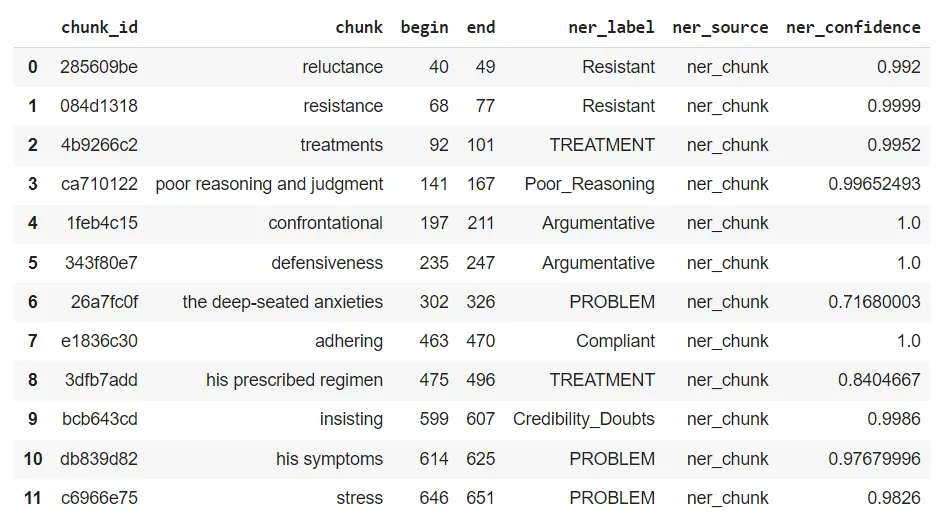
In this study, the developed model successfully accurately identified specific categories of emotions, thoughts, and behaviors within the text. For instance, the model classified the words “reluctance” and “resistance” into the Resistant category with over 99% confidence, indicating that these terms reflect a state of resistance or opposition. Similarly, it labeled the phrases “treatments” and “his prescribed regimen” as Treatment, highlighting themes related to medical treatment and adherence.
The model accurately categorized the phrase “poor reasoning and judgment” as Poor_Reasoning with 99.65% confidence, correctly identifying it as indicative of impaired reasoning and decision-making abilities. Additionally, it classified “confrontational” and “defensiveness” under the Argumentative category, recognizing these words as reflecting argumentative and defensive behavior.
By categorizing phrases like “the deep-seated anxieties,” “his symptoms,” and “stress” as Problem, the model highlighted the underlying issues or symptoms being discussed. Lastly, it identified “adhering” as Compliantand “insisting” as Credibility_Doubts, effectively detecting significant emotional and cognitive states such as compliance and doubts about credibility within the text.
These findings demonstrate the model’s ability to analyze text and accurately classify different emotional and behavioral states with high precision.
Let’s analyze a few more examples:
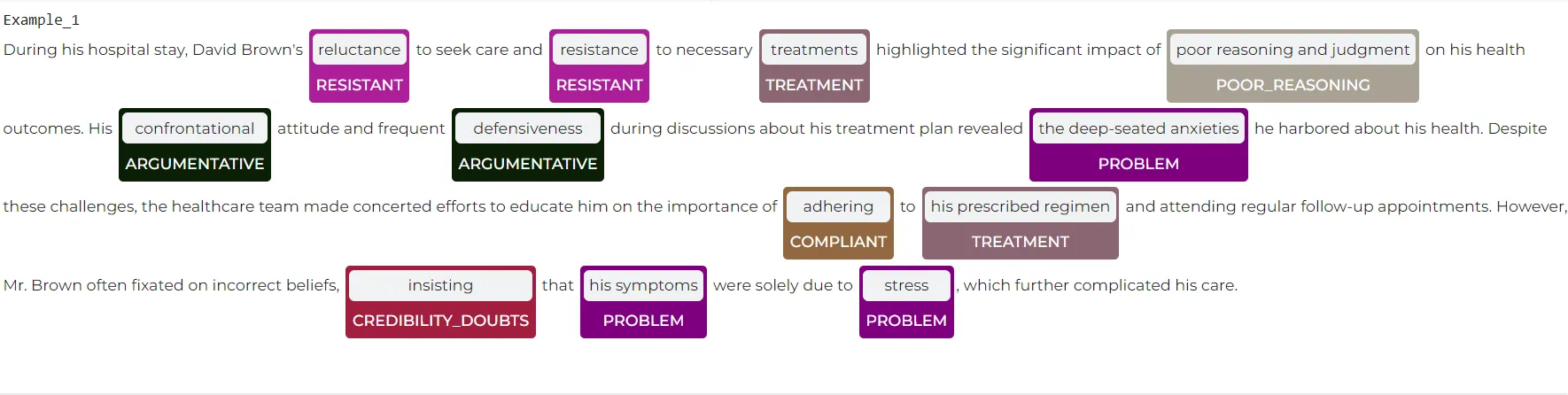
sample_texts = ["""The healthcare team observed that Mr. Smith exhibited somewhat aggressive behavior and was highly irritable, especially when discussing his treatment plan. He showed a full range of emotions and fixated on certain incorrect beliefs about his health. Concerns about his poor insight and judgment were frequently discussed in multidisciplinary team meetings. For example, he often insisted that his symptoms were purely due to stress."""] # Result: +-------------------------+-----+---+-----------------------------+ |chunk |begin|end|ner_label | +-------------------------+-----+---+-----------------------------+ |aggressive |63 |72 |Aggressive | |irritable |98 |106|Aggressive | |poor insight and judgment|269 |293|Poor_Reasoning | |discussed |311 |319|Collaborative_Decision_Making| |insisted |379 |386|Credibility_Doubts | |his symptoms |393 |404|PROBLEM | |stress |425 |430|PROBLEM | +-------------------------+-----+---+-----------------------------+
sample_texts = ["""History of Present Illness: Ms. ___ is a very pleasant ___ female who underwent a left partial mastectomy and left axillary sentinel node biopsy on ___ for left invasive ductal carcinoma. Her surgical pathology report indicated that all six margins were either involved with or close to atypical or carcinoma cells. We decided to go with a global re-excision lumpectomy, which was then performed on ___."""] # Result: +----------------------------------+-----+---+-----------------------------+ |chunk |begin|end|ner_label | +----------------------------------+-----+---+-----------------------------+ |pleasant |46 |53 |Positive_Descriptors | |a left partial mastectomy |80 |104|TREATMENT | |left axillary sentinel node biopsy|110 |143|TEST | |left invasive ductal carcinoma |156 |185|PROBLEM | |atypical or carcinoma cells |287 |313|PROBLEM | |decided |319 |325|Collaborative_Decision_Making| |a global re-excision lumpectomy |338 |368|TREATMENT | +----------------------------------+-----+---+-----------------------------+
Benefits of the Stigmatization NER Model:
- Improved Patient Care: By identifying and reducing stigmatizing language, healthcare providers can foster more respectful and supportive interactions with patients.
- Enhanced Communication: Promotes the use of neutral and person-first language, improving clarity and understanding among medical professionals.
- Bias Reduction: Helps uncover and address implicit biases within medical documentation, contributing to more equitable healthcare practices.
- Compliance and Standards: Assists healthcare organizations in meeting ethical and professional standards regarding patient communication and documentation.
- Educational Tool: Serves as a resource for training healthcare professionals about the impact of language and the importance of mindful communication.
For more information, you can visit ner_stigmatization_wip model.
Conclusion
The language used within medical settings holds immense power in shaping patient experiences and health outcomes. Stigmatizing language can undermine the quality of care, perpetuate health inequalities, and damage the essential trust between patients and healthcare providers. The Stigmatization NER Model developed by John Snow Labs offers a robust solution to identify and address such language within medical records systematically.
By leveraging advanced NLP technologies like Spark NLP, healthcare organizations can proactively enhance their communication practices, ensuring that all patients are treated with dignity and respect. This not only aligns with the ethical obligations of medical professionals but also contributes to more effective and equitable healthcare systems. Embracing tools and models that promote mindful and compassionate language is a critical step toward improving patient care and fostering healthier communities.
Resources
[1] Characterization of Stigmatizing Language in Medical Records v1.0.0 https://physionet.org/content/stigmatizing-language/1.0.0/[2] [PDF] Characterization of Stigmatizing Language in Medical Records https://aclanthology.org/2023.acl-short.28.pdf