Leveraging TextMatcherInternal for Precise Phrase Matching in Healthcare Texts
The TextMatcherInternal annotator in Healthcare NLP is a powerful tool for exact phrase matching in healthcare text analysis. We’ll cover its key parameters and demonstrate its implementation through a practical example. Additionally, we’ll explain how to save and load the model for future use. This annotator can significantly enhance tasks such as clinical document analysis and patient data management.
In the ever-evolving field of healthcare, accurate text analysis can significantly enhance data-driven decisions and patient outcomes. One of the powerful tools in Spark NLP is the TextMatcherInternal annotator, designed to match exact phrases in documents. In addition to the variety of Named Entity Recognition (NER) models available in our Models Hub, such as the Healthcare NLP MedicalNerModel utilizing Bidirectional LSTM-CNN architecture and BertForTokenClassification, our library also features robust rule-based annotators including ContextualParser, RegexMatcher, EntityRulerInternal, and TextMatcherInternal. In this blog post, you’ll learn how to use this annotator effectively in your healthcare NLP projects.
What is TextMatcherInternal?
The TextMatcherInternal annotator is a versatile tool that matches exact phrases provided in an external file against a document. It offers several configurable parameters, allowing you to tailor its functionality to specific use cases.
Parameters:
- setEntities: Sets the external resource for the entities.
path : str
Path to the external resource
read_as : str, optional
How to read the resource, by default ReadAs.TEXT
options : dict, optional
Options for reading the resource, by default {"format": "text"}
- setCaseSensitive: Determines whether the match is case-sensitive (Default: True).
- setMergeOverlapping: Decides whether to merge overlapping matched chunks (Default: False).
- setEntityValue: Sets the value for the entity metadata field if not specified in the file.
- setBuildFromTokens: Indicates whether the annotator should derive the CHUNK from TOKEN.
- setDelimiter: Specifies the delimiter between phrases and entities.
Setting Up Spark NLP Healthcare Library for Enhanced Text Matching
First, you need to set up the Spark NLP Healthcare library. Follow the detailed instructions provided in the official documentation.
Additionally, refer to the Healthcare NLP GitHub repository for sample notebooks demonstrating setup on Google Colab under the “Colab Setup” section.
# Install the johnsnowlabs library to access Spark-OCR and Spark-NLP for Healthcare, Finance, and Legal. ! pip install -q johnsnowlabs
from google.colab import files
print('Please Upload your John Snow Labs License using the button below')
license_keys = files.upload()
from johnsnowlabs import nlp, medical
# After uploading your license run this to install all licensed Python Wheels and pre-download Jars the Spark Session JVM nlp.settings.enforce_versions=True nlp.install(refresh_install=True)
from johnsnowlabs import nlp, medical import pandas as pd
# Automatically load license data and start a session with all jars user has access to spark = nlp.start()
Using TextMatcherInternal for Drug Name Recognition in Clinical Texts
Let’s dive into a practical example using the TextMatcherInternal annotator.
Example: Matching Drug Names in Clinical Text
In this example, we’ll match specific drug names mentioned in clinical text using the TextMatcherInternal annotator.
First, create a source file containing all the chunks or tokens you aim to capture. In the example below, we use # as a delimiter to separate the label from the entity. Therefore, set the parameter as setDelimiter(‘#’). (You can include multiple entities within the same file.)
matcher_drug = """
Aspirin 100mg#Drug
aspirin#Drug
paracetamol#Drug
amoxicillin#Drug
ibuprofen#Drug
lansoprazole#Drug
"""
with open ('matcher_drug.csv', 'w') as f:
f.write(matcher_drug)
Next, define the Spark NLP pipeline:
# Define the Document Assembler
documentAssembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
# Define the Tokenizer
tokenizer = Tokenizer()\
.setInputCols(["document"])\
.setOutputCol("token")
# Define the TextMatcherInternal annotator
entityExtractor = TextMatcherInternal()\
.setInputCols(["document", "token"])\
.setEntities("matcher_drug.csv")\
.setOutputCol("matched_text")\
.setCaseSensitive(False)\
.setDelimiter("#")\
.setMergeOverlapping(True)
# Create the pipeline
mathcer_pipeline = Pipeline().setStages([
documentAssembler,
tokenizer,
entityExtractor])
# Sample data
data = spark.createDataFrame([["John's doctor prescribed aspirin 100mg for his heart condition, along with paracetamol for his fever, amoxicillin for his tonsilitis, ibuprofen for his inflammation, and lansoprazole for his GORD."]]).toDF("text")
# Fit the model and transform the data
matcher_model = mathcer_pipeline.fit(data)
result = matcher_model.transform(data)
# Show results
result.select(F.explode(F.arrays_zip(
result.matched_text.result,
result.matched_text.begin,
result.matched_text.end,
result.matched_text.metadata,)).alias("cols"))\
.select(F.expr("cols['0']").alias("chunk"),
F.expr("cols['1']").alias("begin"),
F.expr("cols['2']").alias("end"),
F.expr("cols['3']['entity']").alias('label')).show(truncate=70)
# Show results
result.select(F.explode(F.arrays_zip(
result.matched_text.result,
result.matched_text.begin,
result.matched_text.end,
result.matched_text.metadata,)).alias("cols"))\
.select(F.expr("cols['0']").alias("chunk"),
F.expr("cols['1']").alias("begin"),
F.expr("cols['2']").alias("end"),
F.expr("cols['3']['entity']").alias('label')).show(truncate=70)
Output:
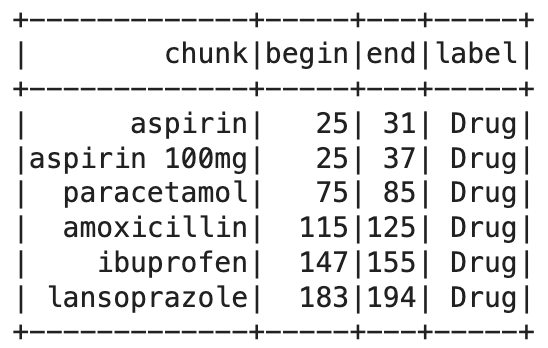
This code creates a CSV file containing drug names and their labels, then sets up a pipeline with a DocumentAssembler, Tokenizer, and TextMatcherInternal. The pipeline processes a sample clinical text, extracting and displaying the matched drug names along with their positions and labels.
TextMatcherInternalModel:
After building and training the TextMatcherInternalmodel, you can save it for future use. Let’s rebuild one of the examples we’ve done before and save it.
entityExtractor = TextMatcherInternal() \
.setInputCols(["document", "token"]) \
.setEntities("matcher_drug.csv") \
.setOutputCol("matched_text")\
.setCaseSensitive(False)\
.setDelimiter("#")
mathcer_pipeline = Pipeline().setStages([
documentAssembler,
tokenizer,
entityExtractor])
data = spark.createDataFrame([["John's doctor prescribed aspirin 100mg for his heart condition, along with paracetamol for his fever and headache, amoxicillin for his tonsilitis, ibuprofen for his inflammation, and lansoprazole for his GORD."]]).toDF("text")
matcher_model = mathcer_pipeline.fit(data)
result = matcher_model.transform(data)
Saving the model:
matcher_model.stages[-1].write().overwrite().save("matcher_internal_model")
Loading and Using the Saved Model
To load and use the saved model, you can use the TextMatcherInternalModel via load.
entity_ruler = TextMatcherInternalModel.load('/content/matcher_internal_model') \
.setInputCols(["document", "token"]) \
.setOutputCol("matched_text")\
.setCaseSensitive(False)\
.setDelimiter("#")
pipeline = Pipeline(stages=[documentAssembler,
tokenizer,
entity_ruler])
pipeline_model = pipeline.fit(data)
result = pipeline_model.transform(data)
This code snippet demonstrates how to save the trained TextMatcherInternal model to disk and load it back for further use. The loaded model is then used in a new pipeline to process clinical text.
Enhancing Healthcare NLP Projects with TextMatcherInternal
The TextMatcherInternal annotator in Spark NLP is a powerful tool for analyzing healthcare texts. It allows for precise and efficient matching of predefined phrases within a given document. By leveraging its flexible parameters, such as case sensitivity and delimiter settings, you can significantly enhance the accuracy and efficiency of your NLP projects. This flexibility makes it ideal for various healthcare applications, from identifying drug names to extracting specific medical conditions.
For more comprehensive examples, detailed documentation, and further insights into utilizing TextMatcherInternal, visit the Spark NLP Workshop.






























