In this notebook, RoBertaForQuestionAnswering was used for versatile Named Entity Recognition (NER) without extensive domain-specific training. This blog post walks through the ZeroShotNerModel implementation and explores its ability to adapt to various datasets quickly and efficiently.
Introduction to ZeroShotNerModel for Healthcare NLP
In the domain of Named Entity Recognition (NER), models traditionally require extensive training on domain-specific data to achieve high accuracy.ZeroShotNerModel, on the other hand, generalizes to a wide range of datasets without requiring additional fine-tuning for every domain.
In addition to the variety of Named Entity Recognition (NER) models available in ourModels Hub, such as the Healthcare NLP MedicalNerModel utilizing Bidirectional LSTM-CNN architecture, BertForTokenClassification, as well as powerful rule-based annotators, our library also includes the versatileZeroShotNerModel for extracting NER.
This blog post will guide you through using theZeroShotNerModel in HealthcareNLP, emphasizing its versatility and practical applications.
What is ZeroShotNerModel?
ZeroShotNerModelis a versatile NER tool designed to recognize entities across different contexts without the need for extensive retraining. This model is particularly useful in scenarios under which one needs to adapt fast to new datasets. As such, it forms a very solid solution toward the execution of different tasks in NER.
Parameters:
entityDefinitions: A dictionary with definitions of the named entities. The keys are entity types, and the values are lists of hypothesis templates.predictionThreshold: The minimal confidence score to consider the entity (Default: 0.01).ignoreEntities: A list of entities to be discarded from the output.
Implementation in Spark NLP
Let’s walk through a practical implementation of ZeroShotNerModel using a sample notebook.
Step 1: Set Up the Environment
First, we need to set up the Spark NLP Healthcare library. Follow the detailed instructions provided in theofficial documentation.
Additionally, refer to the Healthcare NLP GitHub repositoryfor sample notebooks demonstrating setup on Google Colab under the “Colab Setup” section.
# Install the johnsnowlabs library to access Spark-OCR and Spark-NLP for Healthcare, Finance, and Legal. ! pip install -q johnsnowlabs
from google.colab import files print('Please Upload your John Snow Labs License using the button below') license_keys = files.upload()
from johnsnowlabs import nlp, medical
# After uploading your license run this to install all licensed Python Wheels and pre-download Jars the Spark Session JVM
nlp.settings.enforce_versions=True
nlp.install(refresh_install=True)
from johnsnowlabs import nlp, medical import pandas as pd
# Automatically load license data and start a session with all jars user has access to
spark = nlp.start()
Step 2: Defining Unannotated Text to Identify Medical Entities
text_list = [
"The doctor pescribed Majezik for my severe headache.",
"The patient was admitted to the hospital for his colon cancer.",
"27 years old patient was admitted to clinic on Sep 1st by Dr. X for a right-sided pleural effusion for thoracentesis.",
]
data = spark.createDataFrame(text_list, nlp.StringType()).toDF("text")
This data will be used to demonstrate how the ZeroShotNerModel can accurately identify and classify relevant entities like medical conditions, treatments, and patient demographics directly from unannotated text, without the need for specific training data.
Step 3: Defining Pipeline
The ZeroShotNerModel utilizes a pretrained language model (zero_shot_ner_roberta) to perform Named Entity Recognition (NER) on medical text. It identifies entities like medical conditions, drugs, admission dates, and patient ages based on predefined prompts, without needing specific training data for each entity type.
documentAssembler = DocumentAssembler() \
.setInputCol("text") \
.setOutputCol("document")
sentenceDetector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence")
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")
# Zero Shot NER Model
zero_shot_ner = ZeroShotNerModel.pretrained(
"zero_shot_ner_roberta", "en", "clinical/models"
) \
.setEntityDefinitions({
"PROBLEM": [
"What is the disease?",
"What is his symptom?",
"What is her disease?",
"What is his disease?",
"What is the problem?",
"What does a patient suffer",
"What was the reason that the patient is admitted to the clinic?",
],
"DRUG": [
"Which drug?",
"Which is the drug?",
"What is the drug?",
"Which drug does he use?",
"Which drug does she use?",
"Which drug do I use?",
"Which drug is prescribed for a symptom?",
],
"ADMISSION_DATE": ["When did patient admitted to a clinic?"],
"PATIENT_AGE": [
"How old is the patient?",
"What is the age of the patient?",
],
}) \
.setInputCols(["sentence", "token"]) \
.setOutputCol("zero_shot_ner") \
.setPredictionThreshold(0.1)
ner_converter = NerConverterInternal() \
.setInputCols(["sentence", "token", "zero_shot_ner"]) \
.setOutputCol("ner_chunk")
pipeline = nlp.Pipeline(stages=[
documentAssembler,
sentenceDetector,
tokenizer,
zero_shot_ner,
ner_converter,
])
# Fit the pipeline on an empty DataFrame
zero_shot_ner_model = pipeline.fit(spark.createDataFrame([[""]]).toDF("text"))
Results:
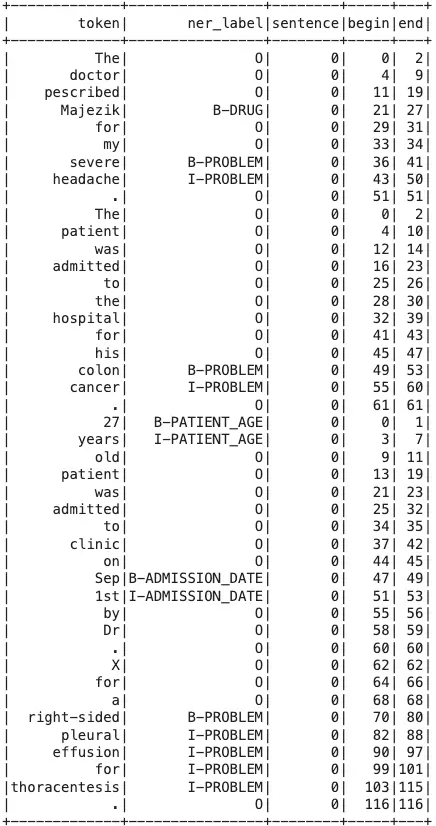
results = zero_shot_ner_model.transform(data) results.show()
Output:

from pyspark.sql import functions as F
results.select(
F.explode(
F.arrays_zip(
results.token.result,
results.zero_shot_ner.result,
results.zero_shot_ner.metadata,
results.zero_shot_ner.begin,
results.zero_shot_ner.end,
)
).alias("cols")
).select(
F.expr("cols['0']").alias("token"),
F.expr("cols['1']").alias("ner_label"),
F.expr("cols['2']['sentence']").alias("sentence"),
F.expr("cols['3']").alias("begin"),
F.expr("cols['4']").alias("end"),
F.expr("cols['2']['confidence']").alias("confidence"),
).show(
50, truncate=100
)
Output:
Conclusion
ZeroShotNerModelin Spark NLP offers a powerful solution for generalized NER tasks, enabling quick adaptation to new datasets. Its ability to generalize without domain-specific training makes it an invaluable tool for various industries, particularly in healthcare where data diversity is prevalent. For more examples and detailed usage, refer to the Spark NLP Workshop.
In this sense, withZeroShotNerModel, data processing workflows can be significantly improved to enable efficient and precise recognition of entities across various datasets.































