Significance for Cancer Diagnosis
Biomarkers (short for biological marker) are measurable biological indicators that provide crucial information about health status, disease processes, or treatment responses. Biomarkers can be molecules, genes, proteins, or other biological characteristics found in blood, tissue, or other samples. In cancer care, they play a vital role in diagnosis, prognosis, and treatment selection. Their accurate detection and interpretation enable personalized medicine approaches, allowing clinicians to tailor treatments to individual patients. As technology progresses, biomarkers continue to evolve, promising more precise and effective cancer care.
Accurate biomarker extraction from clinical notes can greatly enhance cancer diagnosis in several ways:
- Early Detection: Many cancer biomarkers appear in clinical notes before a formal diagnosis. Extracting these early indicators allows for faster identification of potential cancer cases.
- Enhanced Diagnosis And Prognosis: Biomarkers extracted from clinical notes can provide valuable information for more accurate cancer diagnosis and prognosis prediction. This allows clinicians to better understand the specific characteristics of a patient’s cancer.
- Personalized Screening: Biomarker information helps guide the selection of targeted therapies and personalized treatment plans. Certain biomarkers can indicate likely response to specific treatments, allowing for more informed decision-making.
- Research And Discovery: Analyzing biomarker data extracted from large volumes of clinical notes can uncover new correlations and insights, potentially leading to the identification of novel biomarkers or combinations with diagnostic or prognostic value.
- Monitoring Treatment Effectiveness: Tracking biomarker levels mentioned in follow-up notes provides insights into treatment efficacy and disease progression over time.
Biomarker extraction from clinical notes can greatly enhance personalized cancer care and expedite research, ultimately improving patient outcomes and deepening our understanding of cancer biology and treatment.
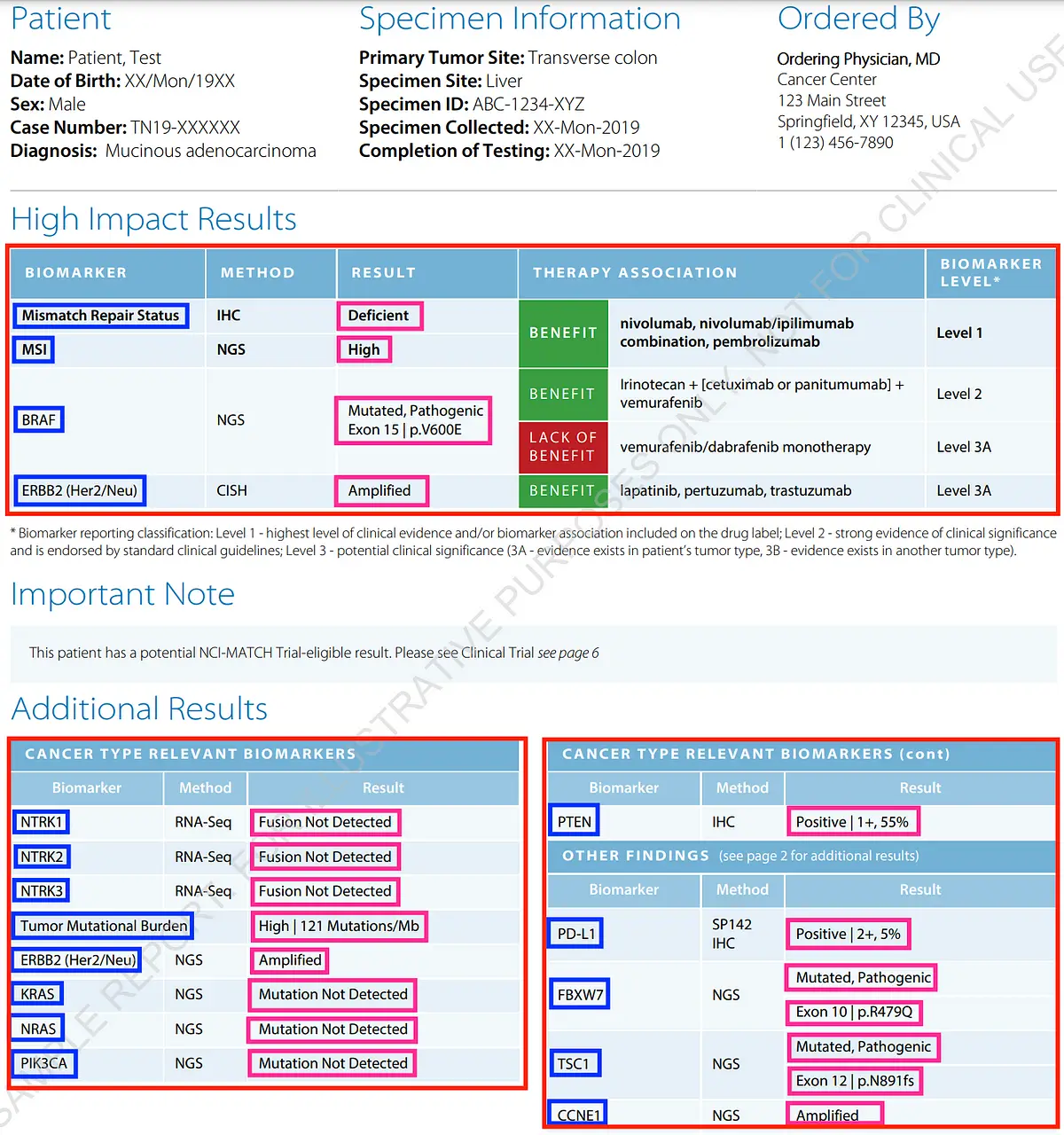
Biomarker And Biomarker Result Table (image resource: Caris Molecular Intelligence-MI Profile Sample Report)
Challenges of Working with Unstructured Clinical Data
Clinical notes are unstructured text datasets, which lack a predefined structure or format and pose significant challenges for data analysis and processing. Here are some of the key difficulties:
- Inconsistent Terminology: Variability in how clinicians document information, including abbreviations and jargon, makes data standardization difficult.
- Context Ambiguity: Understanding the context is challenging, as similar terms and abbreviations can have different meanings based on clinical scenarios.
- Data Quality Issues: Clinical notes often contain errors, incomplete information, or non-standardized text, affecting extraction accuracy.
- Complex Language Structures: Free-text data includes complex syntax, nested information, and nuanced expressions that are hard for algorithms to interpret correctly.
By addressing these challenges, accurate biomarker extraction from clinical notes has the potential to advance personalized cancer care and accelerate cancer research significantly.
Utilizing Healthcare NLP to Detect Cancer Biomarkers in Clinical Notes
Healthcare NLP is a specialized library developed by John Snow Labs, built on the Apache Spark framework so allowing for distributed processing of large volumes of clinical text data, and offers pre-trained clinical pipelines and models for various Natural Language Processing (NLP) tasks in healthcare. These models, trained on medical notes, can accurately extract entities from even messy or unstructured clinical datasets.
We can leverage the Healthcare NLP library, which offers 7700+ different NER models and several pretrained pipelines to assist with clinical entity detection and various NLP tasks in clinical notes, including cancer biomarker detection. Specifically for oncology-related tasks, the library provides 55+ specialized models trained for entity extraction, assertion status detection, relation extraction, and text classification.
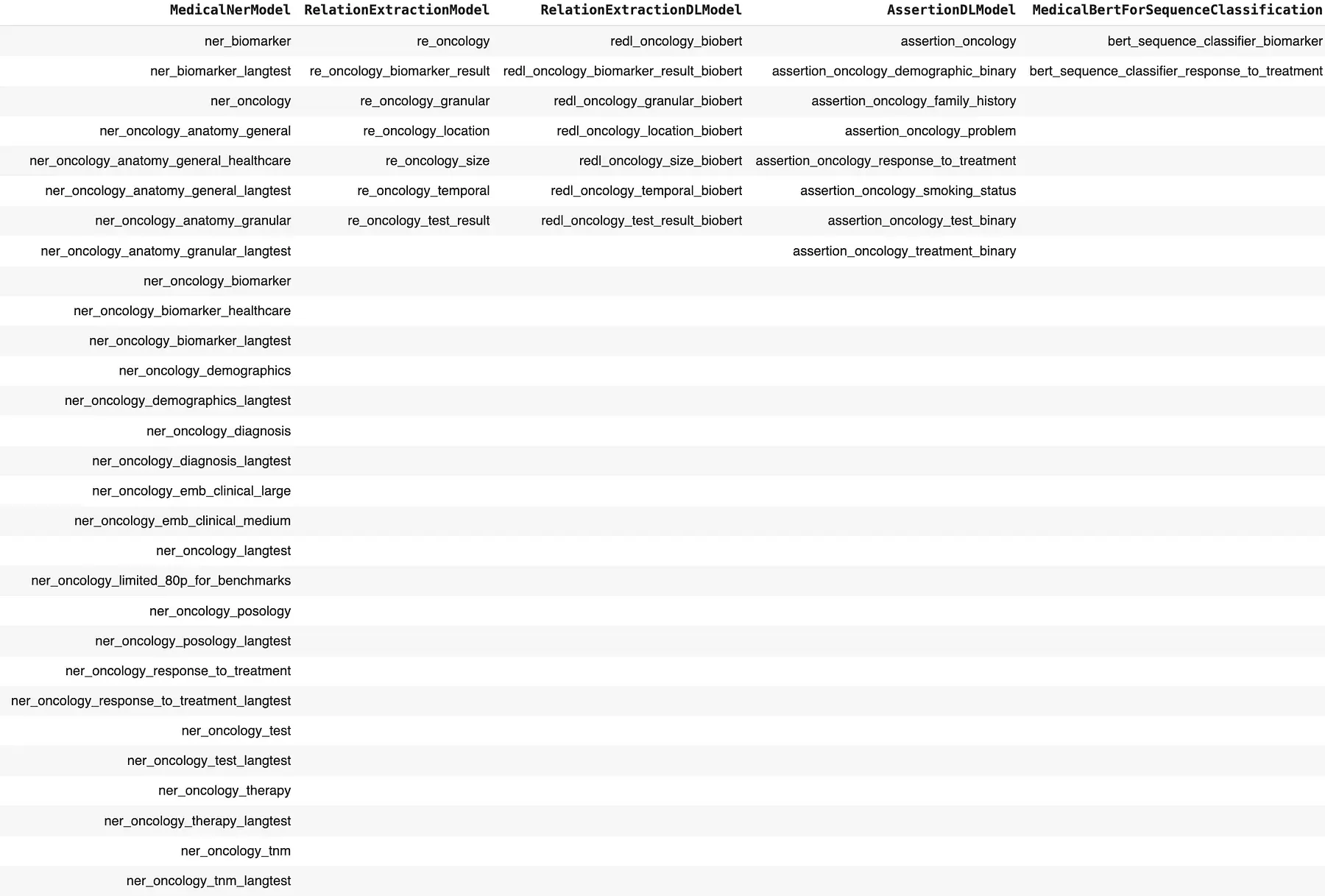
Models For Oncology-Related Tasks
Biomarker and Biomarker Result Table Generation from Oncology Notes
Let’s assume that we are working as an oncology researcher and require a method to efficiently extract and organize biomarker and biomarker result information specifically from sections within oncology notes dedicated to biomarker analysis. This information is crucial for data analysis and biomarker research. Currently, manually extracting this data from extensive oncology notes is time-consuming, labor-intensive, and susceptible to human error.
We can solve this problem by following the solution steps below.
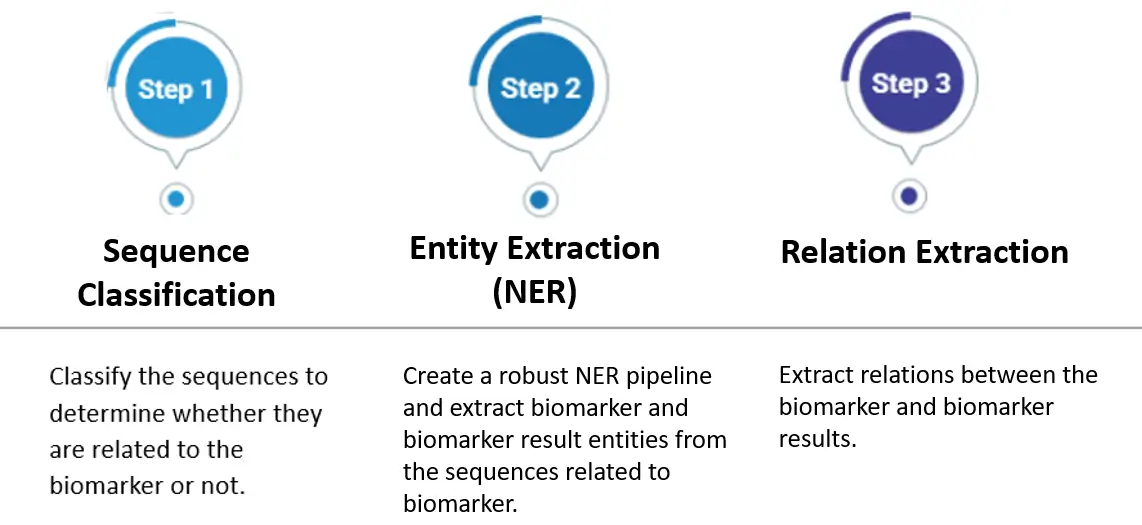
Solution Steps
Sequence Clasification
Sequence classification will assist us in two key ways:
- Some sections of clinical notes may include biomarker entities but are unrelated to biomarkers themselves. By classifying sentences, we can filter out only those that are relevant to biomarkers.
- Certain terms, like “ALK,” can be biomarkers but may also have alternative meanings in clinical notes (e.g., “ALK” as an abbreviation for “Alkaline”). In such cases, the NER model might mislabel these terms. Sequence classification will help identify sentences that genuinely reference biomarkers, reducing misclassification.
- Filtering the sentences will enhance runtime efficiency by eliminating the need to process irrelevant terms in subsequent steps.
For sentence classification, we can use the bert_sequence_classifier_biomarker model to determine whether sentences contain biomarkers.
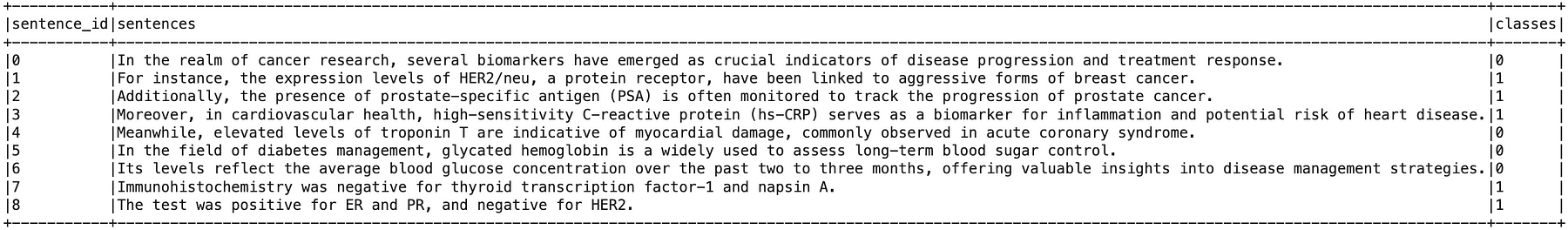
Sentence Classifier Model Results (A value of 1 indicates the presence of a biomarker)
Once classified, we need to filter out the relevant sentences and pass those containing biomarkers to the next stages. The Healthcare NLP library’s DocumentFiltererByClassifier annotator can be used within the pipeline to filter these sentences without any external processing.

Filtered Sentences By DocumentFiltererByClassifier
Entity Extraction
After filtering the sentences, we need to extract the biomarker entities from them. We can create a robust NER pipeline to extract all the biomarker and biomarker result entities from the documents using the severals options in the Healthcare NLP library. To keep it clear, we just used ner_oncology_biomarker model here, but you can add any NER models that can extract biomarker and biomarker result entities.
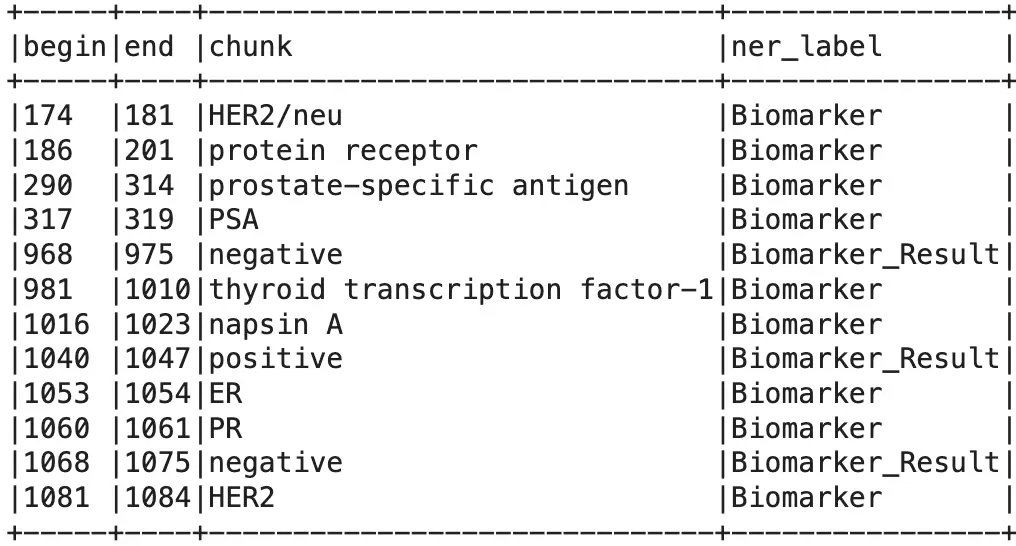
Extracted Biomarker & Biomarker_Result Entities
Relation Extraction
We now have the biomarker and biomarker result entities extracted from the specific sections of the clinical notes. To check their relationship, we can use the re_oncology_biomarker_result relation extraction model, which links biomarker entities to their corresponding results. Alternatively, we can opt for the BERT-based deep learning version of this model, redl_oncology_biomarker_result_biobert.
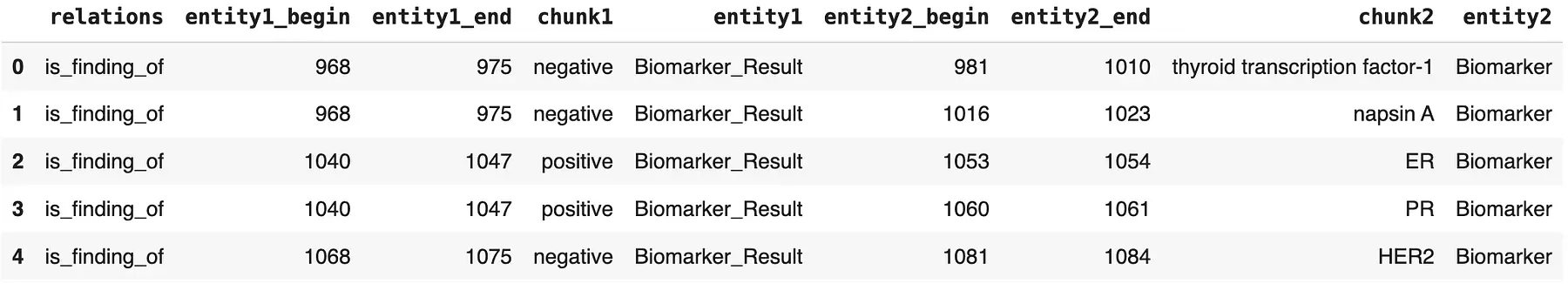
Organized Biomarkers & Biomarker Results
To see how these results can be achieved, you can refer to the complete code in the Oncology Use Case notebook.
Conclusion
Biomarker entity detection from clinical documents plays a crucial role in personalized medicine by enabling more tailored cancer treatments based on individual molecular profiles. It helps guide treatment selection, monitors treatment response, and provides prognostic and predictive information. Biomarkers also aid in matching patients to clinical trials and advancing research by uncovering trends and novel biomarkers. Automated extraction from unstructured clinical notes improves efficiency, supports clinical decision-making, and enhances early cancer detection. Overall, accurate biomarker extraction significantly improves patient outcomes and deepens our understanding of cancer biology and treatment.
The Healthcare NLP library enhances the extraction of biomarker and biomarker result entities by leveraging advanced NLP techniques specifically designed for medical text analysis. Its pre-trained models for named entity recognition identify relevant biomarkers within unstructured clinical notes, while assertion status detection clarifies the context of these biomarkers. Relation extraction further links biomarker entities to their corresponding results, providing deeper insights into their clinical significance. Additionally, entity resolution standardizes these extracted entities, and text classification helps filter out irrelevant data, making the entire process efficient and accurate for personalized medicine and clinical decision-making.
Healthcare NLP models are licensed, so if you want to use these models, you can watch “Get a Free License For John Snow Labs NLP Libraries” video and request one from the John Snow Labs web page.
You can follow us on medium and Linkedin to get further updates or join slack support channel to get instant technical support from the developers of Healthcare NLP. If you want to learn more about the library and start coding right away, please check our certification training notebooks.